Translate this page into:
A preliminary study on the association of single nucleotide polymorphisms of interleukin 4 (IL4), IL13, IL4 receptor alpha (IL4Rα) & Toll-like receptor 4 (TLR4) genes with asthma in Indian adults
Reprint requests: Dr N.B. Ramachandra, Genomics Laboratory, Department of Studies in Zoology, University of Mysore, Manasagangothri, Mysore 570 006, Karnataka, India e-mail: nallurbr@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution NonCommercial ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Interleukin 4 (IL4) and IL13 genes are believed to be responsible for inflammation of the airways in asthmatics. These share a common receptor component called IL4Rα which is another potentially important candidate gene linked to asthma phenotypes. Another gene Toll-like receptor 4 (TLR4) might affect the incidence or progression of asthma through the expression of proinflammatory genes. Several single nucleotide polymorphisms (SNPs) in IL4, IL13, IL4Rα and TLR4 have been reported to be linked to asthma or related phenotypes in several ethnic populations using linkage studies and association studies. However, the results have not been consistent. We investigated five SNPs (C-589T and C-33T of IL4, G+2044A of IL13, A+1902G of IL4Rα, and A+896G of TLR4) in patients with adult onset asthma to evaluate their role in manifestation and severity of asthma.
Methods:
Adult (>18 yr of age) patients with asthma (n=100) and healthy controls (n=50) were included in the study. Genotyping was performed using sequenom MassARRAY technology.
Results:
The mutant alleles of the C-589T and C-33T SNPs in the promoter region of IL4 were present in 4 per cent patients with asthma but absent from the control group suggesting that the variations in IL4 may contribute to asthma occurrence. The SNPs of other genes were seen in both controls and patients.
Interpretation & conclusions:
The results suggest the possible association between the genetic distribution of C-589T and C-33T SNPs of IL4 with asthma in Indian adults.
Keywords
Allele
asthma
cytokine
genes
interleukin
SNPs
TLR4
Asthma is a complex disease characterized by reversible airway obstruction and chronic airway inflammation and is associated with a number of intermediate phenotypes such as elevation of the total serum IgE and airway hyper-responsiveness1. Multiple studies have been carried out to investigate the causes and the risk factors to developing asthma. It has been suggested that interactions among multiple genes and environmental factors increase asthma susceptibility. A variety of environmental factors such as allergens, airborne pollutants, tobacco smokes, and viruses are shown to influence the development of allergic diseases including asthma2.
Genome-wide linkage studies and candidate gene approaches have been used to identify asthma susceptibility genes. Several loci linked to asthma or related phenotypes have been reported using genome-wide linkage studies34. Human chromosome 5q31-33 is among the regions that has shown linkage to asthma5. This region contains cytokine cluster and harbour genes for the T-helper Type 2 (Th2) cytokines such as interleukin 4 (IL4) and IL13. Both these cytokines have been implicated in the pathogenesis of asthma and their associations with asthma and atopy have been reported67. Two single nucleotide polymorphisms (SNPs) in the promoter region of IL4 (C-589T and C-33T) and one variant in the fourth exon of IL13 (G+2044A) have been identified in relation to asthma phenotypes78910. IL4 and IL13 also share a receptor component, the α chain of the IL4R (IL4Rα), which is an essential component of both the IL4 and the IL13 signal transduction pathway. The IL4Rα is located on chromosome 16q12 and is another potentially important candidate gene linked to asthma11. Several IL4R polymorphisms have been shown to be associated with a higher risk of atopic asthma. The Q576R variant (G1902A) in exon 12 of the IL4Rα was found to be associated with a higher risk of atopy, atopic asthma and variation in IgE levels12.
Toll-like receptor 4 (TLR4) belongs to the TLR family and is a part of the endotoxin receptor complex which recognizes endotoxin and activates the innate immune system through the expression of proinflammatory genes13. TLR function might be involved in the development of asthma phenotypes. An A896G polymorphism in the fourth exon of the TLR4 is shown to alter the extracellular domain of this receptor and confers hyporesponsiveness to endotoxin in human14. The association studies which have been carried out in several ethnic groups to find out the associations of these polymorphisms with asthma or related phenotype have not shown consistent results. In this study, we analyzed five SNPs (C-589T and C-33T of IL4, G+2044A of IL13, A+1902G of IL4Rα, and A+896G of TLR4) to determine the involvement of these SNPs in the manifestation and severity of asthma in Indian adults.
Material & Methods
Subjects: The study included 100 adult patients with asthma aged more than 18 yr and 50 non asthmatic controls. The asthma patients were selected consecutively from patients attending a tertiary care asthma center (Allergy, Asthma, and Chest Center, Mysore, India), during 2009-2010. Asthma in the index adult was diagnosed according to Global Initiative for Asthma (GlNA) guidelines15 with reversible airway obstruction of 12 per cent and 200 ml improvement in forced expiratory volume in 1 sec (FEV1) after inhalation of salbutamol. Spirometry was performed according to American Thoracic Society Guidelines16. Patients were categorized based on Global Initiative for Asthma (GINA) guidelines15 to different asthma severity groups that included 15 patients with mild asthma, 30 patients with moderate and 55 patients with severe asthma. Non asthmatic controls had no history of asthma and were selected randomly from the general population of Mysore. The patients and controls who had other chronic respiratory symptoms were excluded from the study.
The study was approved by the Institutional Human Ethical Committee (IHEC) of the University of Mysore, and informed written consent was obtained from all cases and controls who participated in this study.
SNP genotyping: Five SNPs were selected for this study; two SNPs in the promoter region of IL4 (C-589T and C-33T), one SNP in IL13 (G+2044A), one SNP in IL4Rα (A+1902G), and one SNP in the TLR4 (A+896G). The SNP details and sequence data were obtained from NCBI database using the unique accession numbers (www.ncbi.nlm.nih. gov). Genomic DNA was extracted from blood using the DNA isolation kit for mammalian blood (Roche, USA, catalogue number: 11667327001, version October 2008) following the manufacturers’ instructions. Genotyping of the five SNPs was performed using sequenom MassARRAY technology (Sequenom®, San Diego, CA, USA) at Vimta Labs Ltd in Hyderabad, India. It consisted of Sequenom-iPLEX® Gold SNP genotyping platform with SpectroCHIP® and MALDI-Time of Flight (TOF) Mass spectrometer.
Statistical analysis: Genotypes were tested for Hardy-Weinberg equilibrium in patients and controls separately. Differences in the distribution of genotypes and alleles between groups were estimated by univariate statistical analysis (Chi-square test) on SPSS 18.0. (SPSS Inc., Chicago, IL, USA).
Results & Discussion
The demographic profile of the patients and controls is presented in Table I. Schematic representation of the IL4, IL13, IL4Rα and TLR4 showing their structural organization and the location of the genotyped SNPs are shown in Fig. 1.
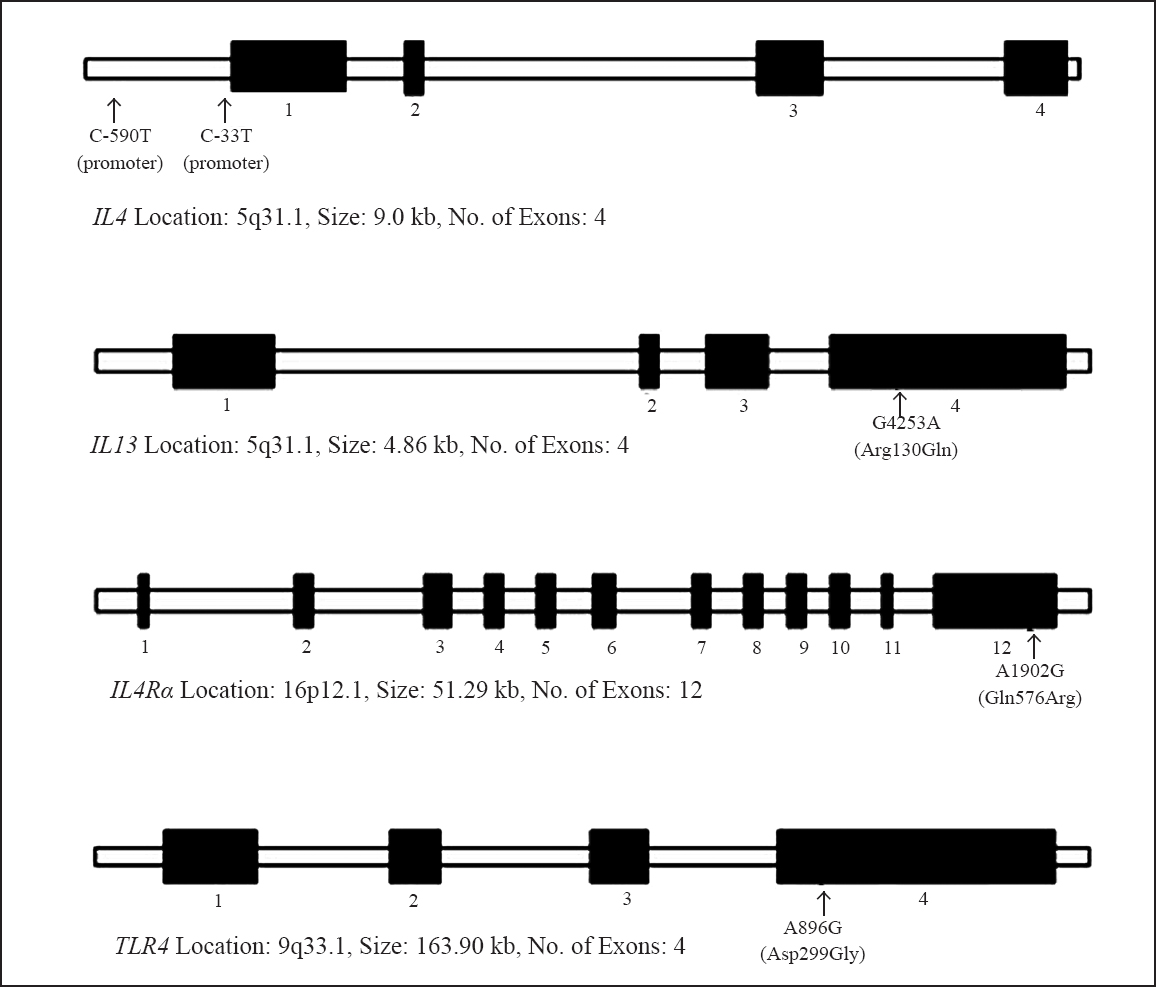
- Schematic representation of interleukin 4 (IL4), IL13, IL4Rα and Toll-like receptor 4 genes (TLR4) showing their structural organization (filled boxes represent the exons) and the location of the SNPs genotyped in this study.
In this study, the homozygous mutant genotype (TT) of both the -589T and -33T SNPs in the promoter region of IL4 was present in four per cent patients with asthma. These two mutations were absent from the control group (Table II). The normal genotype (CC) of C589T SNP was seen in 65 per cent of the patients and 72 per cent of the controls. The heterozygous genotype (CT) of C-589T SNP was present in 31 per cent patients and 28 per cent controls. In case of C-33T SNP in the proximal promoter region of IL4, the normal genotype (CC) was seen in 68 per cent of the patients and 78 per cent of controls. The heterozygous genotype (CT) of C-33T SNP did not show a significant difference between the asthmatics and the controls (Table II).
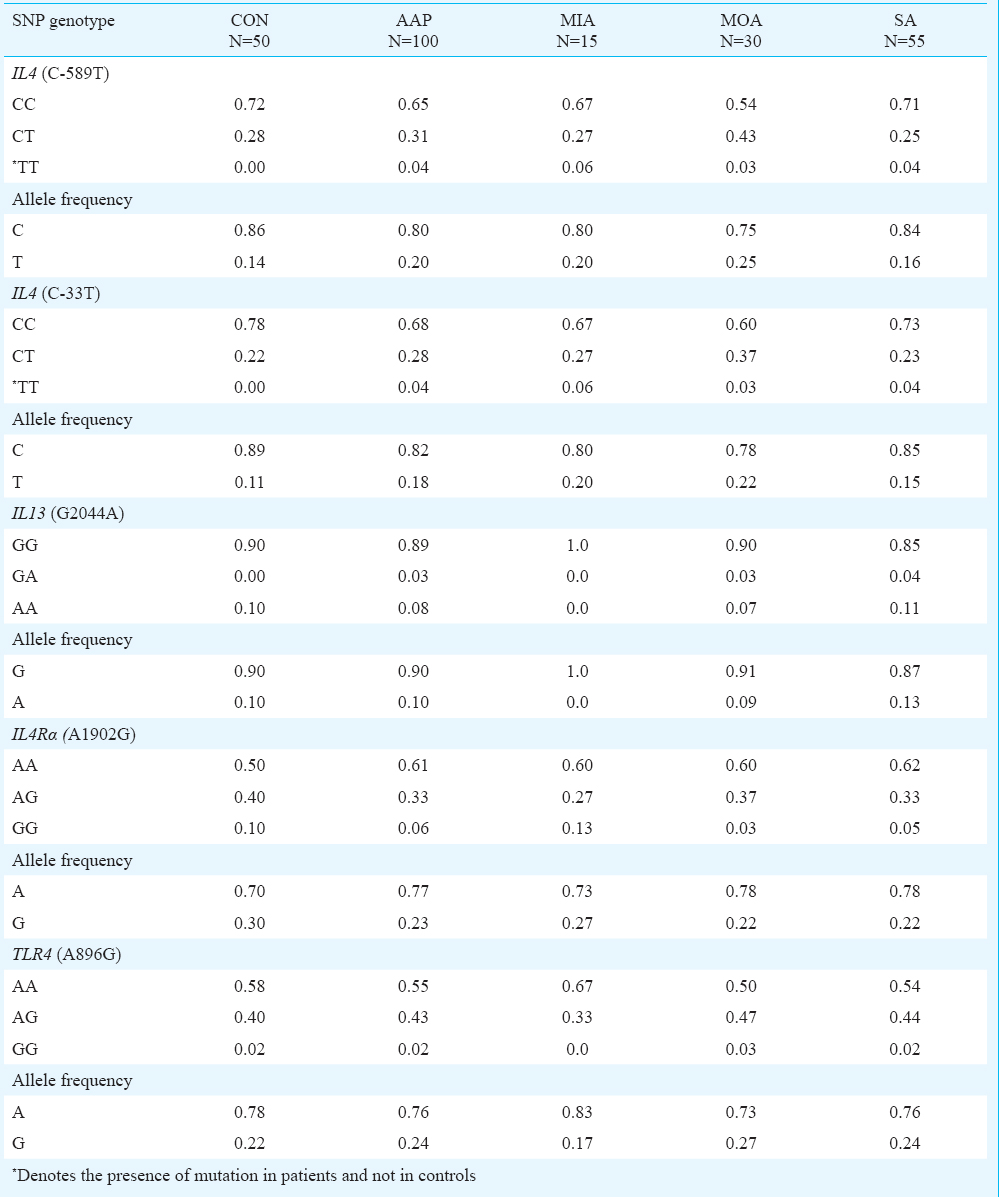
The SNP genotype and allele frequencies of C-589T, C-33T, G+2044A, A+1902G and A+896G in patients and controls are provided in Table II. No significant differences of genotypes were observed in G+2044A, A+1902G and A+896G SNPs. Dominant alleles were more frequent than the recessive alleles in all the five SNPs of the four genes in this study (Table II). After comparing the genotype frequency distribution of all the SNPs between each subgroup of asthma severity and the controls, no significant differences was observed. The intergroup comparisons of genotype frequency of the five SNPs in the three subgroups of asthma severity also did not show any significant differences (data not shown). Distribution in percentage of normal homozygous, heterozygous and mutant homozygous genotypes of the five SNPs in mild, moderate and severe asthma is shown in Fig. 2.
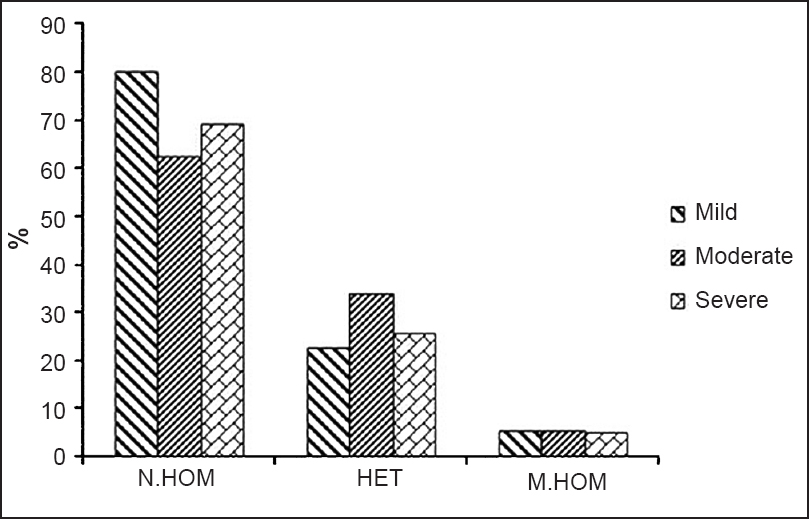
- Distribution of normal homozygous (N.HOM), heterozygous (HET) and mutant homozygous (M.HOM) genotypes of five SNPs of IL4, IL4R, IL13 and TLR4 in mild, moderate and severe asthmatic patients
We had previously measured the serum concentrations of IL13 and interferon gamma (IFN-γ) for the same group of patients17. Increased serum concentration of IL13 was observed in the homozygous mutant genotype of the five SNPs. Also in the control group, the homozygous recessive genotype of IL13 showed a dramatic increase in its serum concentration (data not shown). No significant effect was seen between different genotypes of the five SNPs and the serum concentration of IFN-γ.
In our previous study IL4 C-589T SNP was investigated in Mysore, where higher allele frequency of the homozygous mutant genotype (TT) of -589T IL4 was reported in patients with severe asthma (7%) compared to that of the controls (2%), but the difference was not significant18. The interesting result of this study was the presence of the homozygous mutant allele (T allele) of both C-589T and C-33T SNPs in the promoter region of IL4 in 4 per cent of the patients. Both the mutations were absent in the control group. Transcription of IL4 has been shown to be regulated by multiple promoter elements. It has also been reported that polymorphisms in the promoter region of IL4 correlates with enhanced IL4 activity that is higher binding of transcription factors and increased transcription19.
The limitations of this study were small sample size and selection of the patients from a tertiary care centre. Therefore, the results may not be reflective of the general population. Therefore, it is important to replicate this study in a larger sample size. Taken together, our data suggest the possible association between the genetic distribution of C-589T and C-33T SNPs of IL4 with asthma in Indian adults. It would however, appear to exclude a role for Arg130Gln variation in IL13 and Q551R variation in IL4Rα in the overall susceptibility to asthma. This study also confirms the previous observed lack of association of TLR4 polymorphism (Asp299Gly) with asthma20. Further studies in larger populations should examine the role of these polymorphisms in the development and severity of asthma phenotypes, in order to generalize these results.
References
- An understanding of the genetic basis of asthma. Indian J Med Res. 2011;134:149-61.
- [Google Scholar]
- Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126:453-65.
- [Google Scholar]
- A genome-wide search for quantitative trait loci underlying asthma. Nature. 1996;383:247-50.
- [Google Scholar]
- A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics. 1999;58:1-8.
- [Google Scholar]
- Meta-analysis for linkage to asthma and atopy in the chromosome 5q31-33 candidate; region. Hum Mol Genet. 2001;10:891-9.
- [Google Scholar]
- IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269-74.
- [Google Scholar]
- A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506-13.
- [Google Scholar]
- Relationship between polymorphisms in IL4 and asthma in Japanese women: the Kyushu Okinawa Maternal and Child Health Study. J Investig Allergol Clin Immunol. 2013;23:242-7.
- [Google Scholar]
- Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol. 2000;106:135-40.
- [Google Scholar]
- Interleukin-4 (IL4) and interleukin-4 receptor (IL4RA) polymorphisms in asthma: a case control study. Clin Mol Allergy. 2005;29:3-15.
- [Google Scholar]
- IL4Rá mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Resp Crit Care Med. 2007;175:570-6.
- [Google Scholar]
- Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66:517-26.
- [Google Scholar]
- Innate immune responses to microbial poisons: discovery and function of the Toll-like receptors. Annu Rev Pharmacol Toxicol. 2003;43:609-28.
- [Google Scholar]
- TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187-91.
- [Google Scholar]
- Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008;31:143-78.
- [Google Scholar]
- Serum levels of interleukin-13 and interferon-gamma from adult patients with asthma in Mysore. Cytokine. 2012;60:431-7.
- [Google Scholar]
- Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415-22.
- [Google Scholar]
- Regulation of interleukin 4 gene transcription: alterations in atopic disease? Am J Respir Crit Care Med. 2000;162:S81-5.
- [Google Scholar]
- Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci. 2008;114:347-60.
- [Google Scholar]






