Translate this page into:
A novel reassortant avian influenza H4N6 virus isolated from an environmental sample during a surveillance in Maharashtra, India
For correspondence: Dr Shailesh D. Pawar, Avian Influenza Group, ICMR-National Institute of Virology, 130/1, Sus Road, Pashan, Pune 411 021, Maharashtra, India e-mail: shaileshpawarniv@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Low pathogenic avian influenza (LPAI) viruses cause mild clinical illness in domestic birds. Migratory birds are a known reservoir for all subtypes of avian influenza (AI) viruses. The objective of the study was to characterize AI H4N6 virus isolated from an environmental sample during surveillance in Maharashtra, India.
Methods:
AI surveillance in wild migratory birds was conducted during the winter migratory bird season (2016-2017) in Pune, India. AI H4N6 virus was isolated from the faecal droppings of a wild migratory waterbird. Virological and molecular characterization of the isolated virus was carried out. Virus titration, haemagglutination inhibition assay, receptor specificity assay, intravenous pathogenicity index and neuraminidase inhibition assays were performed. Full genome sequencing, molecular and phylogenetic analyses were also conducted.
Results:
The virus was found to be of low pathogenicity, with avian type receptor specificity, and was susceptible to neuraminidase inhibitors. Phylogenetic and molecular analysis revealed that the present virus is a result of extensive reassortment with AI H8N4, H6N2, H4N3 and H3N6, predominantly as donor viruses among others.
Interpretation & conclusions:
This is the first report of the isolation and characterization of an LPAI H4N6 virus from an environmental sample from India. The present study showed that the H4N6 virus is a novel reassortant and divergent as compared with the reported H4N6 viruses from poultry in India, indicating independent introduction. This highlights the role of wild and migratory birds in the transmission of AI viruses and necessity of such studies at the human-animal interface.
Keywords
Avian influenza
birds
environment
H4N6
phylogeny reassortment
Avian influenza (AI) viruses are a highly heterogeneous group with varying pathogenicity in different species1. AI viruses are classified into two forms based on the severity of the illness caused in poultry. Low pathogenic AI (LPAI) viruses cause mild clinical symptoms whereas highly pathogenic AI (HPAI) viruses cause severe respiratory illness and death among infected chickens1. The global circulation of AI viruses is a major concern due to their ability to acquire high pathogenicity from low-pathogenic strains and/or reassortment with each other to form novel AI viruses with varying pathogenicity2. AI viruses can also emerge as HPAI viruses from an LPAI virus even in the absence of reassortment3.
Besides their natural host reservoirs such as wild birds, waterfowl, gulls and shorebirds, AI viruses also circulate in poultry4. In addition to posing a threat to human health, LPAI outbreaks lead to severe economic losses to the poultry industry. There have been several reports of the LPAI H4N6 from poultry and wild ducks across the world567. Genetically, divergent reassortant H4N6 viruses have been reported as a result of intermixing among different AI subtypes8. Isolations of AI H4N6 and other mammalian adapted H4 viruses of the avian lineage from pigs have been reported from the USA, China and Canada9. These viruses possessed substitutions in the HA protein which enabled adaptation to mammalian type sialic acid receptors (alpha-2,6-linked sialic acid). There have been speculations about the potential for cross-species transmission of H4 viruses on the basis of two amino acid substitutions each in HA and PB2 proteins10. There have also been reports of the seroprevalence of antibodies against AI H4 subtypes in poultry workers11. Liang et al12 reported that in addition to possessing dual receptor binding specificity, AI H4 viruses also possessed limited airborne transmissibility, suggesting the potential threat posed to public health.
In India, apart from outbreaks of HPAI, LPAI viruses such as H4N6, H9N2 and H11N1 in poultry and wild migratory birds have also been reported131415. Wild migratory birds congregate at different water bodies, and several sites have been recognized for their arrival. The winter migratory bird season in India is from October to March. The first report of LPAI H4N6 in India was in the year 2010 from domestic chickens and ducks15, however, not from wild birds. So far, there are no reports of the isolation of an H4N6 virus from environmental samples from India. The present study reports the isolation, virology and molecular characterization of a reassortant AI H4N6 virus from India.
Material & Methods
Collection of specimens and virus isolation: AI surveillance was undertaken during the winter migratory bird season spanning from December 2016 to March 2017 in Pune district, Maharashtra, India. Water bodies such as backwater of dams, lake and reservoirs which are known to host wild and migratory birds. The surveillance sites having water bodies included Bhigwan, Kumbhargaon, Dhumalwadi, Pimpri and Lonand (18.1758° N 74.4516° E, 18.2677° N 74.7864° E, 17.8756° N 74.4767° E, 18.6298° N 73.7997° E and 18.0417° N 74.1862° E, respectively). A total of 222 environmental samples (213 droppings + nine water samples) were collected in Viral Transport Medium (VTM; 2 mL) containing antibiotics (penicillin - 2000 U/mL, streptomycin - 0.2 mg/mL, gentamycin - 0.25 mg/mL). pH of the droppings was recorded using pH indicator strips. The samples were transported to the laboratory in cold-chain. They were processed further in the laboratory and all the samples were inoculated in ten-day-old embryonated white-leghorn clean chicken eggs (Venkateshwara Hatcheries Ltd., Pune) as per the protocol mentioned previously16. Embryonated chicken eggs are preferred for virus isolation as most of the AI viruses grow readily in these17. Briefly, the inoculated eggs were incubated at 37°C for 72 h with daily observation. The allantoic fluids from all the inoculated eggs were harvested irrespective of the mortality, and haemagglutination assay performed using 0.5 per cent Turkey red blood cells as per the WHO protocol17. The allantoic fluids showing HA titre <2 were considered negative.
Haemagglutination and neuraminidase subtyping: For identification of the HA subtype, haemagglutination inhibition assay was performed17 (WHO, 2002). Viral RNA was extracted using QIAamp viral RNA mini kit (Qiagen, Germany) according to manufacturer’s instructions, and one-step reverse transcription-polymerase chain reaction (RT-PCR) was performed using the NA diagnostic primers to identify the NA subtype18.
Virus titration: The 50 per cent egg infectious dose (EID50) titration was performed as per the method mentioned by Klimov et al19. Briefly, ten-fold serial dilutions of the virus isolate were performed in PBS and inoculated in four eggs per dilution. The eggs were incubated for 72 h, allantoic fluids were harvested and haemagglutination assay titres were determined. The EID50 titres were then calculated using the Reed and Muench 50 per cent endpoint determination method.
Sialidase assay: The sialidase assay was performed to determine the receptor-binding specificity of the viruses as described previously20. Briefly, 50 μl of one per cent goose red blood cell suspension prepared in PBS was treated with 1.25 units of alpha 2,3-sialidase enzyme (Takara Bio Inc., Japan) for 1 h at 37°C. These sialidase enzyme-treated RBCs were then used in HA assay using standard protocol17.
Intravenous pathogenicity index: The Intravenous Pathogenicity Index (IVPI) was determined. Briefly, 5-6-wk-old white-leg-horn chickens (n=10) were infected by intravenous route with 0.2 ml of 1:10 diluted virus allantoic fluid containing 103.33 EID50 virus. The chickens were housed in an animal isolator (Montair Process Technology, The Netherlands). The control chickens (n=2) received 0.2 ml PBS and were housed separately. Chickens were observed daily for clinical signs and symptoms for 10 days. Faecal droppings were collected and were processed for virus isolation in 10-day-old embryonated chicken eggs. Intravenous pathogenicity index was determined as described in the WHO Manual on Animal Influenza Diagnosis and Surveillance17. All experiments were approved by the Institutional Animal Ethics Committee (IAEC).
Fluorescence-based neuraminidase inhibitor assay: The susceptibility to NA inhibitors oseltamivir and zanamivir was determined using the fluorescence-based neuraminidase inhibition (NAI) assay21. The NAIs of oseltamivir carboxylate and zanamivir were used in the assay. The viruses, namely A/Fukui/20/2004 wild type H3N2 and A/Fukui/45/2004 (H3N2) 119V mutant (kindly provided by the WHO Collaborating Center for Reference and Research on Influenza, Melbourne, Australia) were used as reference standards for susceptibility and resistance, respectively, since no other reference viruses were available for the NA subtype N6. The plots and the IC50 values were calculated for the sample and standards by using the curve fitting software JASPER (CDC, USA).
One-step reverse transcription-polymerase chain reaction (RT-PCR) and full genome sequencing: All the eight gene segments were amplified by one-step RT-PCR using SuperScript III Platinum one-step RT-PCR system (Invitrogen, USA) reagents and specific primers. Full genome sequencing was carried out using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The sequences were determined using an automated 3130 XL Genetic Analyzer (Applied Biosystems, USA). Nucleotide sequences were assembled and aligned using ‘SeqScape’ software v 2.5.0 (Applied Biosystems, USA) and edited using ‘BioEdit’ (v 7.0.9.1, CDC, Atlanta, USA). The sequences of all the eight gene segments have been submitted to GenBank (accession numbers ‘MK453333 to MK453340’).
Phylogenetic and molecular analysis: Since the isolate belonged to the subtype H4N6, the dataset for gene-wise phylogenetic analysis was assembled by retrieving the available sequences of H4N6 viruses from the Global Initiative on Sharing AI Data (GISAID) EpiFlu database22. Furthermore, to determine the strains which had closest identity with the present isolate BLAST was performed, and sequences with higher percentage nucleotide identities were included in the study (Table I). Multiple sequence alignment and construction of phylogenetic trees were performed using MEGA v5.05. Molecular analysis was performed using ‘BioEdit’ (v 7.0.9.1, CDC, Atlanta, USA).
| Common name | Family | Scientific name | Status (migratory/resident/RM) |
|---|---|---|---|
| Bar-headed goose | Anatidae | Anser indicus | Migratory |
| Ruddy shelduck | Anatidae | Tadorna ferruginea | Migratory |
| Northern shoveller | Anatidae | Anas clypeata | Migratory |
| Indian spot-billed duck | Anatidae | Anas poecilorhyncha | Resident |
| Eurasian wigeon | Anatidae | Anas penelope | Migratory |
| Little egret | Ardeidae | Egretta garzetta | Resident |
| Grey heron | Ardeidae | Ardea cinerea | Resident |
| Cattle egret | Ardeidae | Bubulcus ibis | Resident |
| Intermediate egret | Ardeidae | Egretta intemedia | Resident |
| Red-wattled lapwing | Charadriidae | Vanellus indicus | Resident |
| Painted stork | Ciconiidae | Mycteria leucocephalus | RM |
| Asian openbill | Ciconiidae | Anastomus oscitans | Resident |
| Brown-headed gull | Laridae | Larus brunnicephalus | Migratory |
| Pallas’s gull | Laridae | Larus ichthyaetus | Migratory |
| Western yellow wagtail | Motacilidae | Motacilla flava | Migratory |
| Little cormorant | Phalacrocoracidae | Phalacrocorax niger | Resident |
| Greater flamingo | Phoenicopteridae | Phoenicopterus ruber | RM |
| Black-winged stilt | Recurvirostridae | Himantopus himantopus | RM |
| Little stint | Scolopacidae | Calidris minuta | Migratory |
| Western black-tailed godwit | Scolopacidae | Limosa limosa | Migratory |
| Whiskered tern | Sternidae | Chlidonias hybrida | RM |
| Indian river tern | Sternidae | Sterna aurantia | Resident |
| Glossy ibis | Threskiornithidae | Plegadis falcinellus | Resident |
| Eurasian spoonbill | Threskiornithidae | Platalea leucorodia | RM |
RM, resident migratory
Results
Specimen collection, virus isolation and haemagglutination assay: Among a total of 24 wild bird species which were observed, nine were migratory, ten resident and five resident-migratory bird species (Table II). The pH of the droppings ranged between 6.5 to 8.0. No mortality was observed in any of the inoculated eggs. Out of 222 harvested allantoic fluids tested in HA, virus was isolated from one faecal dropping of a water bird, which showed an HA titre of 32 HAU. All the other allantoic fluids were negative in HA assay.
| Gene | Virus | Accession number | Subtype | PNI |
|---|---|---|---|---|
| PB2 | A/mallard/Chany/126K-2/2014 | EPI1388622 | H8N4 | 98 |
| A/mallard/Chany/126K/2014 | EPI884262 | H5N3 | 98 | |
| A/mallard duck/Netherlands/56/2015 | EPI1306909 | H3N2 | 98 | |
| PB1 | A/duck/Hubei/ZYSYG3/2015 | EPI942215 | H6N2 | 98 |
| A/northern shoveler/Egypt/MB-D-695C/2016 | EPI1581322 | H7N3 | 98 | |
| A/mute swan/Croatia/102/2016 | EPI873617 | H5N5 | 98 | |
| PA | A/duck/Bangladesh/31227/2016 | EPI1099011 | H6N2 | 98 |
| A/black-necked crane/Zhaotong/ZT-12/2013 | EPI1133530 | H1N2 | 98 | |
| A/duck/Hokkaido/X9/2016 | EPI1510517 | H8N4 | 98 | |
| HA | A/duck/Mongolia/17/2011 | EPI1153577 | H4N3 | 96 |
| A/duck/Mongolia/118/2015 | EPI704283 | H4N6 | 95 | |
| A/duck/Bangladesh/25891/2015 | EPI965331 | H4N6 | 93 | |
| NP | A/teal/Egypt/MB-D-487OP/2016 | EPI1581280 | H7N3 | 99 |
| A/teal/Egypt/MB-D-621C/2016 | EPI1581314 | H7N9 | 99 | |
| A/pintail/Egypt/MB-D-384C/2015 | EPI1581278 | H3N6 | 99 | |
| NA | A/mallard/Toguchin/19/2017 | EPI1328510 | H4N6 | 98 |
| A/duck/Sichuan/04.08 CDLQ033-O/2015 | EPI659686 | H4N6 | 97 | |
| A/duck/Mongolia/146/2010 | EPI1153565 | H3N6 | 97 | |
| M | A/gadwall/Chany/315/2016 | EPI925991 | H1N1 | 98 |
| A/mallard/Chany/126K-2/2014 | EPI1388625 | H8N4 | 98 | |
| A/northern shoveler/Egypt/MB-D-695C/2016 | EPI1581326 | H7N3 | 98 | |
| NS | A/duck/Hubei/ZYSYG8/2015 | EPI942277 | H6N2 | 99 |
| A/duck/Mongolia/499/2015 | EPI704567 | H10N7 | 99 | |
| A/duck/Mongolia/278/2011 | EPI1134008 | H3N8 | 99 |
PNI scores obtained from GISAID BLAST. Representative sequences of H4N6 viruses, as well as those of other subtypes sharing maximum identity with H4N6-1722760. Multiple nucleotides and amino acid sequence alignments for all the eight gene segments were performed using MEGA (v5.05). Top three representative hits showing maximum identity have been included. PNI, per cent nucleotide identity, GISAID, Global Initiative on Sharing Avian Influenza Data
Haemagglutinin and neuraminidase subtyping: The virus subtype was identified as H4N6 based on haemagglutination inhibition assay and NA diagnostic RT-PCR. The isolated H4N6 virus was assigned the nomenclature: ‘A/migratory bird/India/1722760/2017 (H4N6)’; hereafter referred to as ‘H4N6-1722760’.
Virus titration, sialidase assay, intravenous pathogenicity index and neuraminidase inhibition assay: EID50 titre of H4N6-1722760 was 104.33/0.2 ml. Sialidase assay revealed avian type receptor specificity. The IVPI score of the infected chickens was 0.00 indicating low-pathogenicity. Faecal droppings of infected chickens were tested at the end of the experiment on day 10 and were positive for virus isolation. H4N6-1722760 was susceptible to both oseltamivir carboxylate and zanamivir with IC50 values 0.71 and 0.03 nM, respectively.
Molecular analysis: The sequences of H4N6-1722760 were analysed for their genetic characteristics. All the gene segments were screened for known molecular markers. There was an absence of multibasic aminoacids at the HA cleavage site, indicating that it was an LPAI virus. The HA sequence had 226Q and 228G (H3 numbering) at the receptor-binding region, implying avian virus-like receptor specificity (sialic acid -2,3-NeuAcGal)23. No amino acid deletion in the NA stalk region was observed. These results corroborated findings of the sialidase assay and intravenous pathogenicity index (IVPI). The avian-associated residues 158E, 271T, 590G, 591Q, 627E and 701D were observed in PB2. The substitutions in the M2 protein which confer amantadine resistance were absent. H4N6-1722760 showed that a full-length PB1-F2 protein of 90 amino acids, wherein N66S mutation, known to increase the virulence of influenza A virus by inhibiting the early interferon response, was present24. The NS1 protein had an alanine (A) residue at position 149, which is known to enable antagonization of interferon induction in chick embryo fibroblast cells25. At the C-terminal of the NS1 protein, the ESEV motif was observed. The presence of this motif is known to cause increased virulence and pathogenicity in experimentally infected mice, similar to that reported in HPAI H5N1 viruses26. However, pathogenicity in the mammalian model was not tested.
The per cent nucleotide identity (PNI) of the isolated strain H4N6-1722760 with representative H4N6 strains isolated from poultry in India in the year 2010 was compared. The PB2, PB1, PA, NP, NA and M genes ranged between 92 and 97 per cent. The PNIs of the HA genes of the three previous H4N6 isolates with H4N6-1722760 were 84 per cent. The PNIs of the NS genes of previous H4N6 isolates with H4N6-1722760 were 74 per cent for chicken isolates and 96 per cent for the duck isolate (Table III).
| Gene | Virus | Accession number | PNI |
|---|---|---|---|
| PB2 | A/Duck/India/10736/2009 | MK453341.1 | 97 |
| A/chicken/India/101006/2009 | MK453347.1 | 93 | |
| A/chicken/India/101018/2009 | MK453354.1 | 93 | |
| PB1 | A/Duck/India/10736/2009 | MK453342.1 | 94 |
| A/chicken/India/101006/2009 | MK453348.1 | 95 | |
| A/chicken/India/101018/2009 | MK453355.1 | 95 | |
| PA | A/Duck/India/10736/2009 | MK453343.1 | 94 |
| A/chicken/India/101006/2009 | MK453349.1 | 96 | |
| A/chicken/India/101018/2009 | MK453356.1 | 96 | |
| HA | A/Duck/India/10736/2009 | JX310059.1 | 84 |
| A/chicken/India/101006/2009 | JX310061.1 | 84 | |
| A/chicken/India/101018/2009 | JX310062.1 | 84 | |
| NP | A/Duck/India/10736/2009 | MK453344.1 | 94 |
| A/chicken/India/101006/2009 | MK453350.1 | 92 | |
| A/chicken/India/101018/2009 | MK453357.1 | 92 | |
| NA | A/Duck/India/10736/2009 | JX310060.1 | 96 |
| A/chicken/India/101006/2009 | JX310063.1 | 96 | |
| A/chicken/India/101018/2009 | JX310064.1 | 95 | |
| M | A/Duck/India/10736/2009 | MK453345.1 | 97 |
| A/chicken/India/101006/2009 | MK453351.1 | 97 | |
| A/chicken/India/101018/2009 | MK453358.1 | 97 | |
| NS | A/Duck/India/10736/2009 | MK453346.1 | 96 |
| A/chicken/India/101006/2009 | MK453352.1 | 74 | |
| A/chicken/India/101018/2009 | MK453359.1 | 74 |
PNI scores obtained using NCBI BLAST. PNI, per cent nucleotide identity; NCBI, National Center for Biotechnology Information
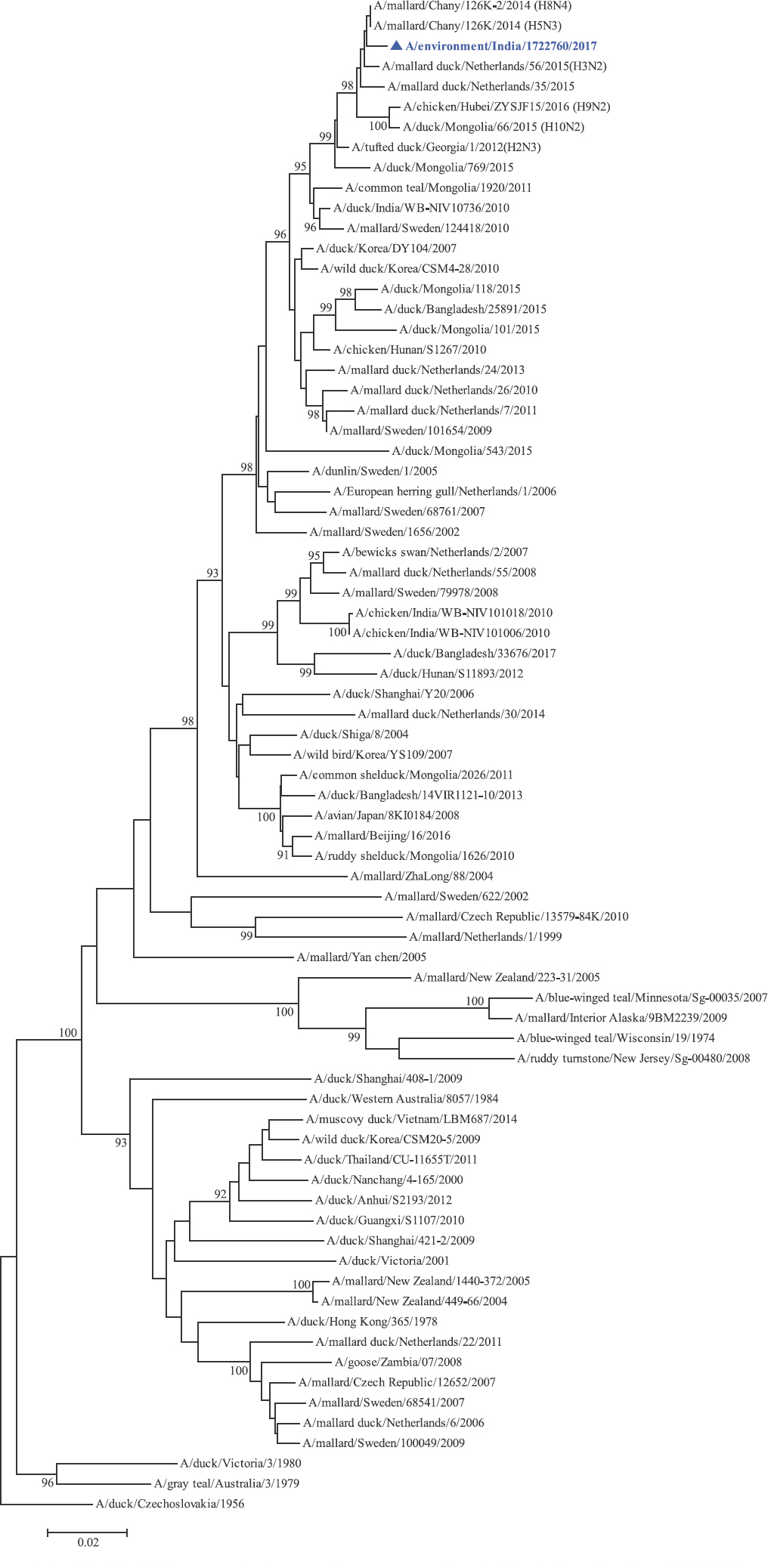
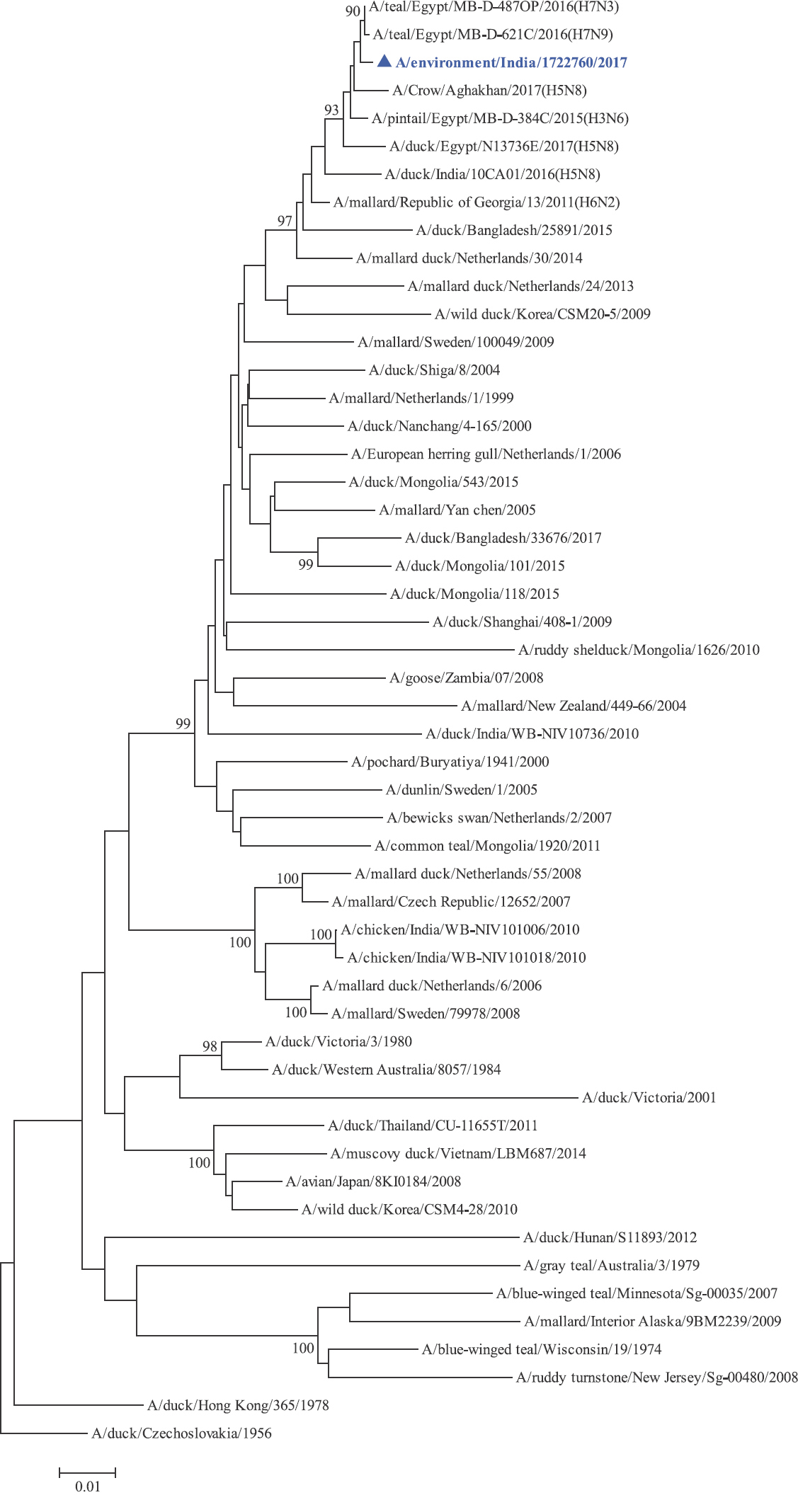
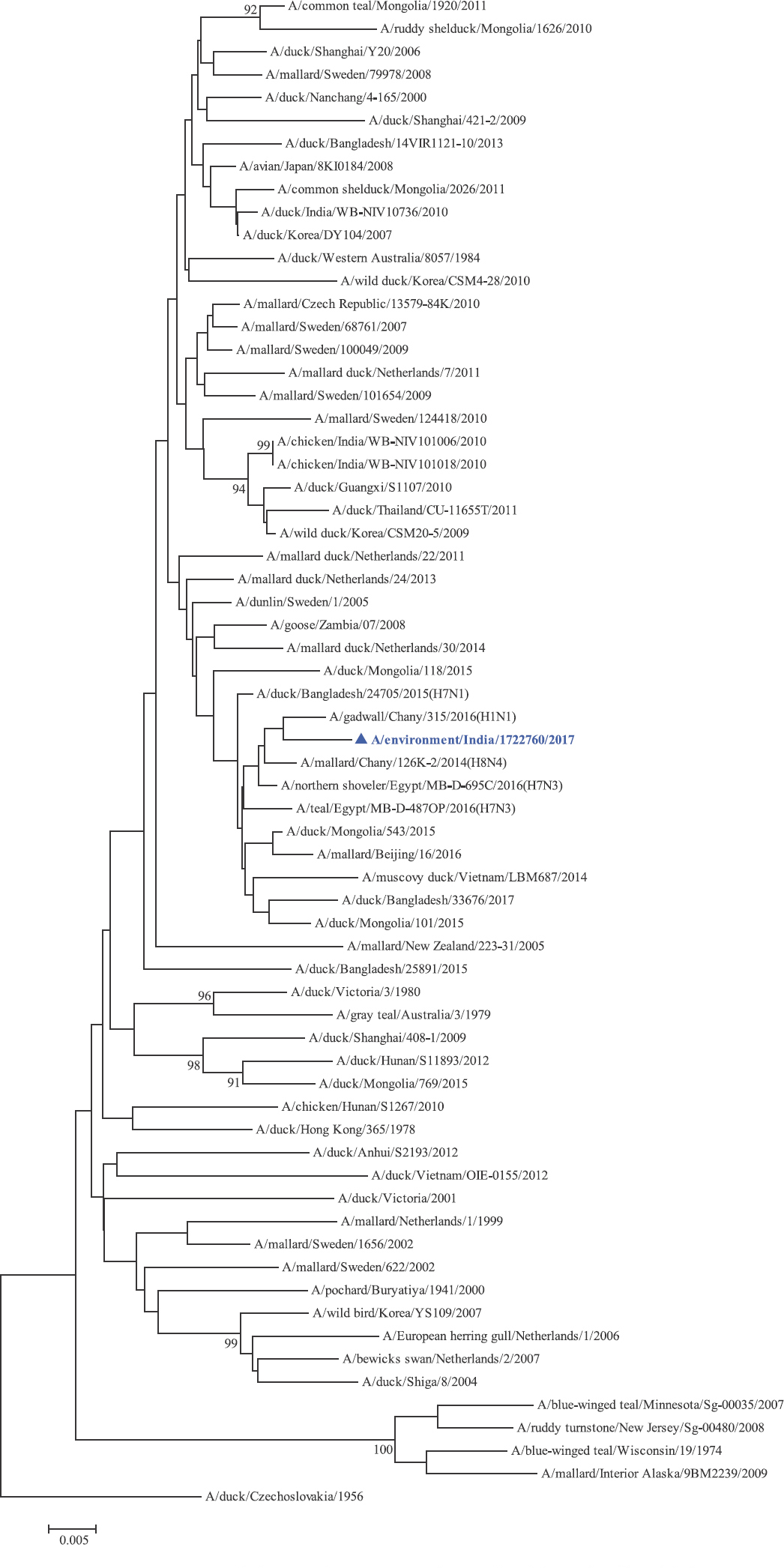
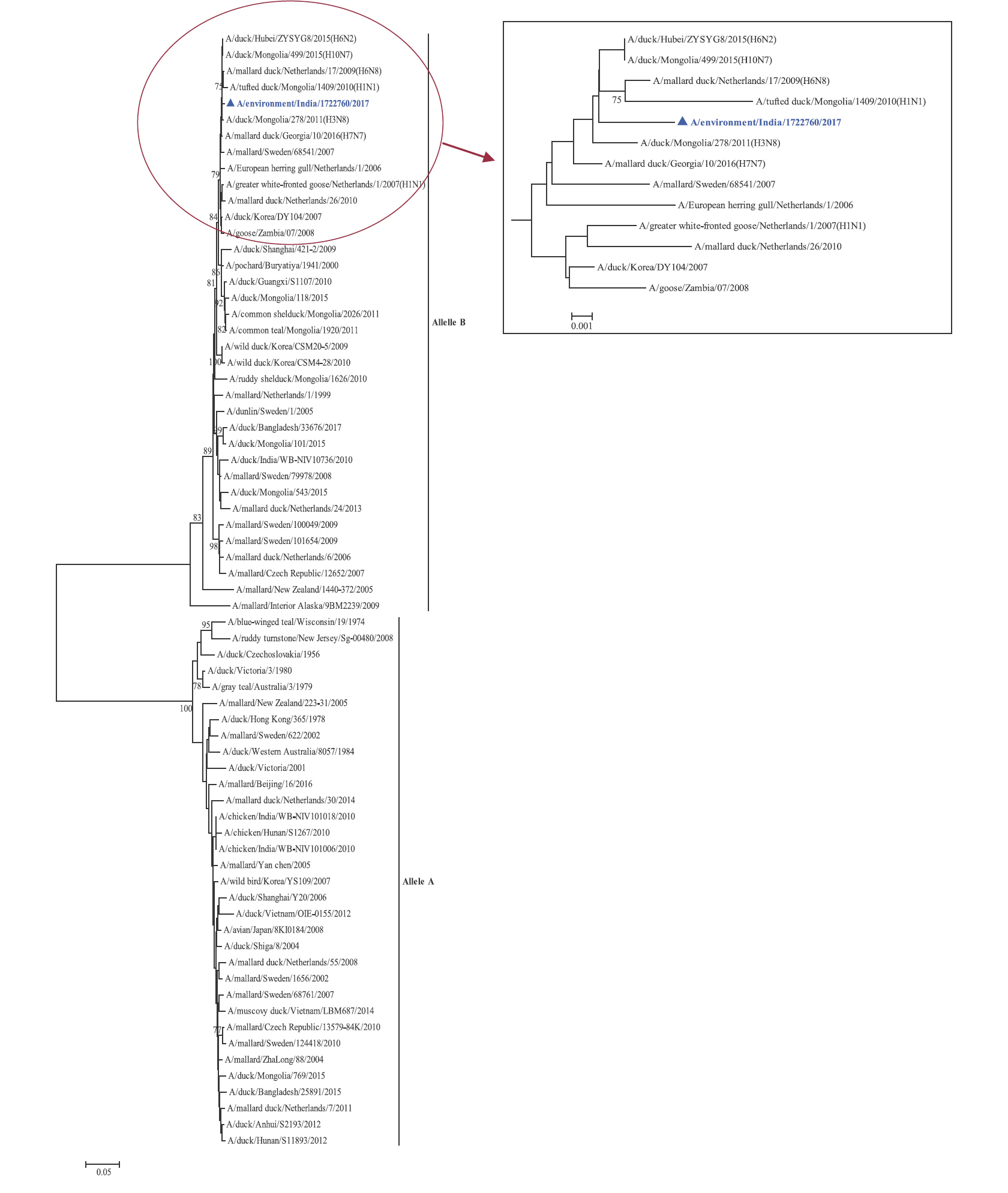
Phylogenetic analysis: Phylogenetic trees for all the eight gene segments revealed that H4N6-1722760 belonged to the Eurasian lineage (Fig. 1). The HA of the present virus showed divergence from the HA genes with the maximum per cent nucleotide identity being 96 per cent for A/duck/Mongolia/17/2011(H4N3); and 95 and 93 per cent for H4N6 viruses A/duck/Mongolia/118/2015 and A/duck/Bangladesh/25891/2015, respectively (Fig. 1A). For the NA gene, H4N6-1722760 bore closest resemblance to N6 viruses from Mongolia, Sichuan province in China and from Toguchin, Russia (Fig. 1B). In the phylogenetic tree of PB2 gene, H4N6-1722760 closely grouped with the PB2 of H8N4 and H5N3 viruses from Chany, Russia and an H3N2 virus from the Netherlands (Fig. 1C), whereas for PB1, the closest isolates were those from Hubei (H6N2), Egypt (H7N3) and an HPAI H5N5 virus from Croatia (H5N5) (Fig. 1D). In the phylogenetic tree of the PA gene, H4N6-1722760 was clustered with the viruses from Bangladesh (H6N2), China (H1N2) and Japan (H8N4) (Fig. 1E). For NP, H4N6-1722760 closely grouped with H7N3 and H7N9 viruses from Egypt and an HPAI H5N8 virus from Aghakhan, Iran (Fig. 1F). For the M gene, H4N6-1722760 was closely related to H8N4 and H1N1 viruses from Chany, Russia and an H7N3 virus from Egypt (Fig. 1G). For the NS gene, H4N6-1722760 grouped in the allele B (Fig. 1H), usually encountered in avian viruses, with resemblance to an H6N2 virus from Hubei, China, and H10N7 and H3N8 viruses from Mongolia. It has been shown experimentally that although its introduction in mammalian hosts is rare, allele B does not attenuate viral replication27. Hence, the possibility of infection in mammals cannot be ruled out.
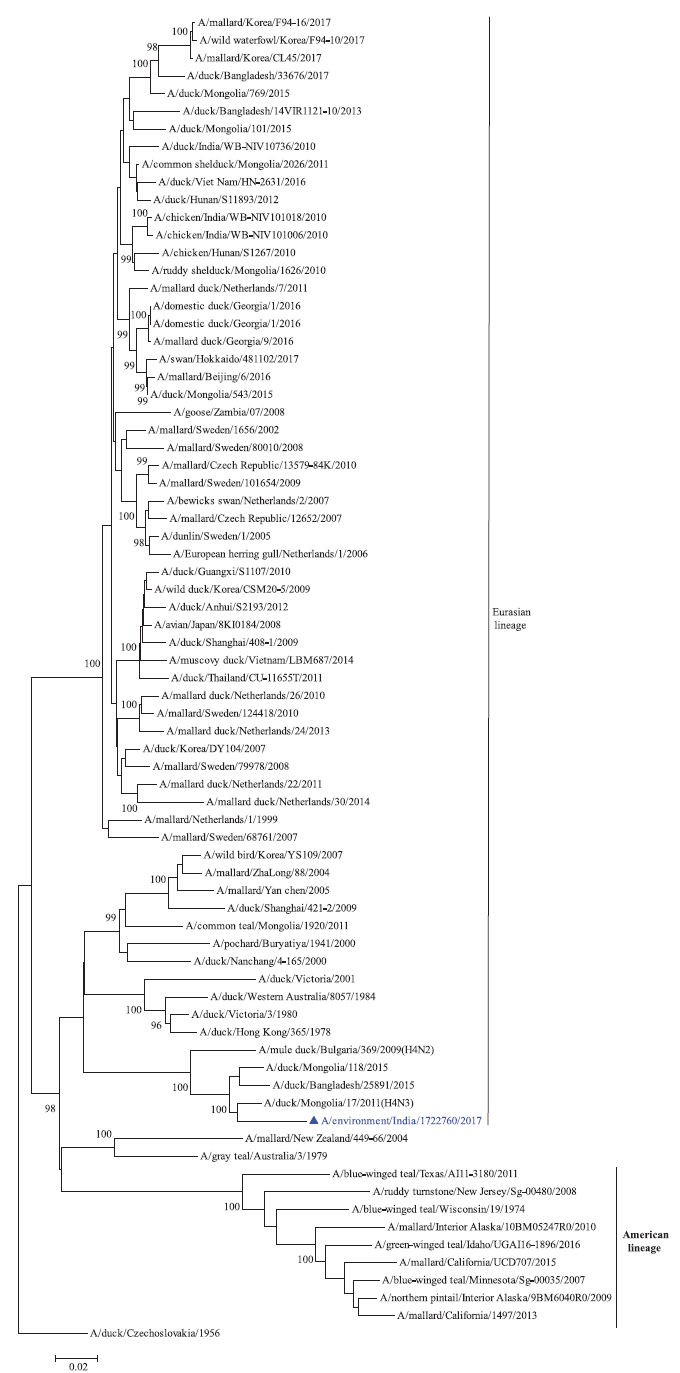
- Phylogenetic trees for all the eight gene segments of H4N6-1722760 constructed using the neighbour-joining method with the Kimura 2-parameter distance model and 1000 bootstrap replicates in MEGA v5.05. A/migratory bird/India/1722760/2017 (H4N6) is shown in blue with a triangle. (A) HA gene phylogenetic tree rooted to the virus A/duck/Czechoslovakia/1956, the Eurasian and American lineages have been labelled.
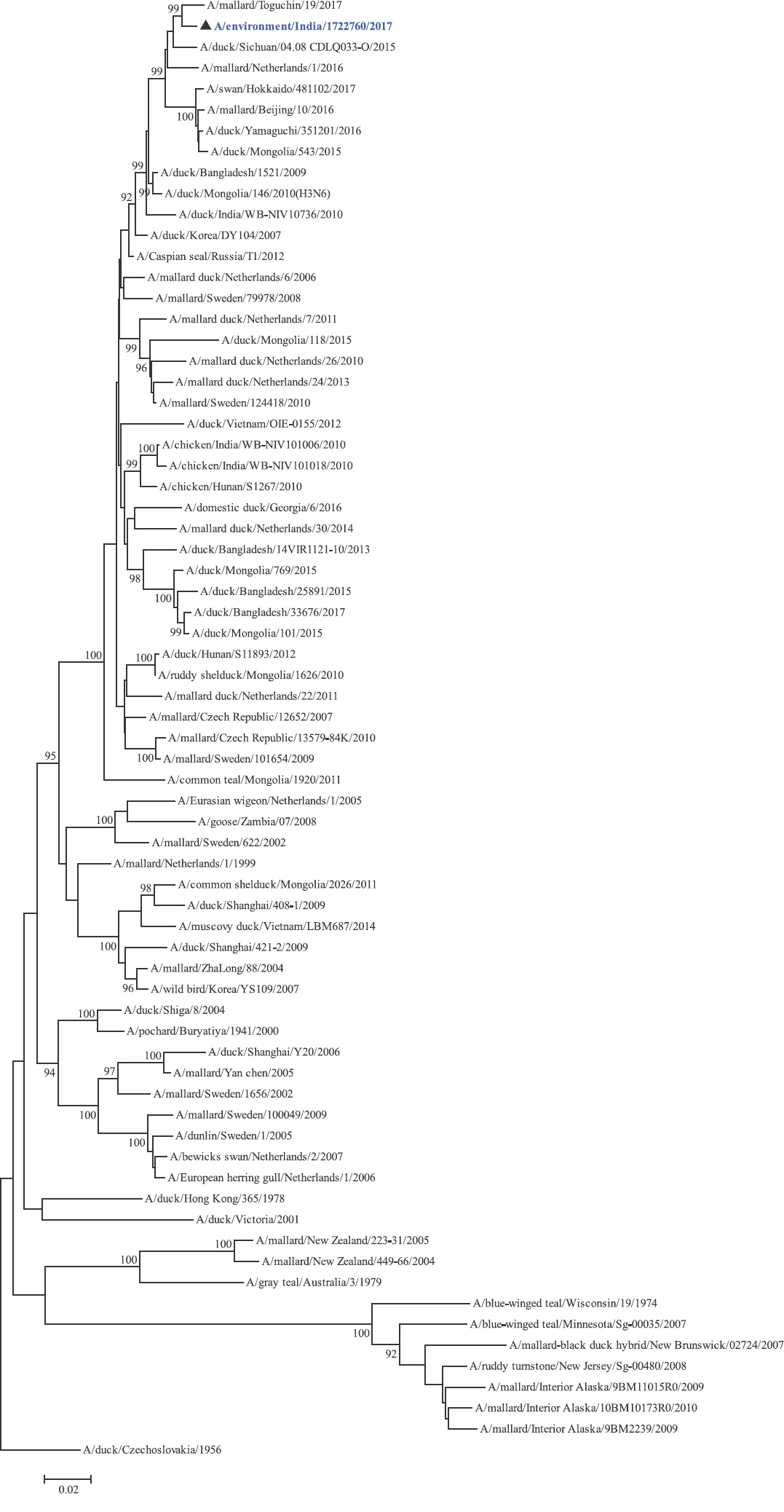
-
NA gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.
-
PB2 gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.
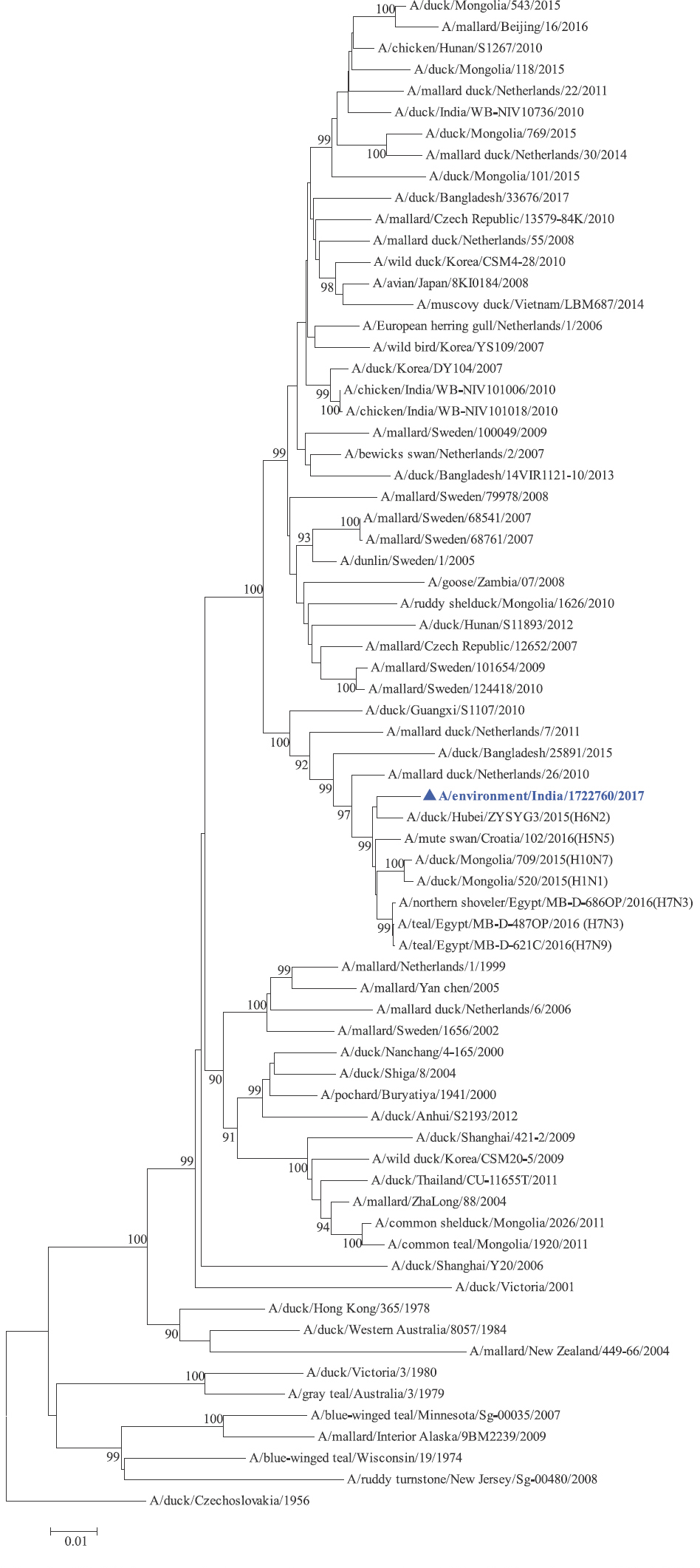
- PB1 gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.

-
PA gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.
-
NP gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.
-
M gene phylogenetic tree rooted to A/duck/Czechoslovakia/1956.
-
NS gene phylogenetic tree, the two alleles A and B have been labelled, the cluster containing A/migratory bird/India/1722760/2017 (H4N6) has been enlarged in the inset.
Phylogenetic analysis and high per cent nucleotide identities with viruses of varied subtypes indicated that the present virus is a result of complex and extensive reassortment events, as illustrated in Fig. 2. The PB2 and M genes of H4N6-1722760 were contributed by A/mallard/Chany/126K-2/2014(H8N4)-like viruses; PB1, PA and NS genes by A/duck/Hubei/ZYSYG3/2015(H6N2)-like; HA gene by A/duck/Mongolia/17/2011(H4N3)-like; NP gene by A/pintail/Egypt/MB-D-384C/2015(H3N6)-like and NA gene by A/duck/Sichuan/04.08 CDLQ033-O/2015(H4N6)-like viruses.
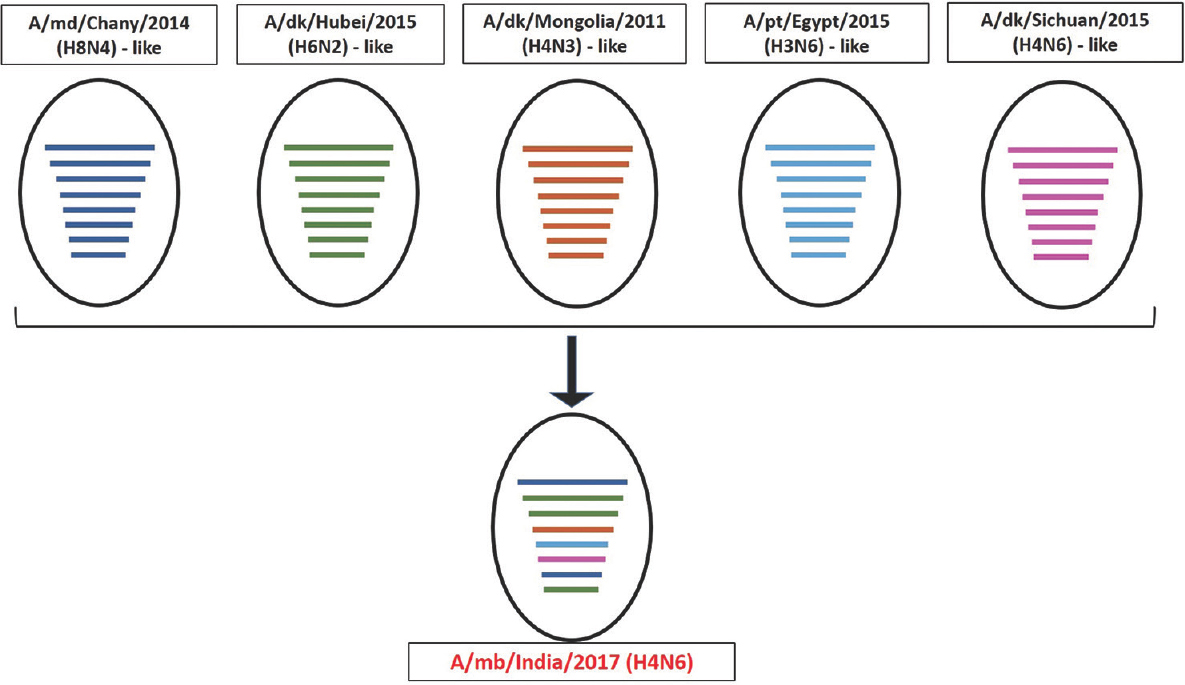
- Schematic diagram for proposed origin of the reassortant H4N6-1722760. The proposed genetic constellation of H4N6-1722760 has been represented schematically, depicting the genesis of the reassortant. The abbreviations md, dk and pt stand for mallard, duck and pintail, respectively. A/mb/India/2017 (A/migratory bird/India/1722760/2017) represents the reassortant virus in the present study. The circles represent virus strains, whereas coloured horizontal lines represent the individual gene segments of the respective viruses (PB2, PB1, PA, HA, NP, NA, MP, NS genes). The virus A/mallard/Chany/126K-2/2014 (H8N4) contributed PB2 and M genes (represented in blue), A/duck/Hubei/ZYSYG3/2015 (H6N2) contributed PB1, PA and NS genes (represented in green), A/duck/Mongolia/17/2011 (H4N3) contributed the HA gene (represented in light brown), A/pintail/Egypt/MB-D-384C/2015 (H3N6), NP gene (represented in sky blue) and A/duck/Sichuan/04.08 CDLQ033-O/2015 (H4N6) contributed the NA gene (represented in pink).
Discussion
So far, there are no reports of isolation of H4N6 virus from the environmental samples from wild migratory birds in India, although LPAI H4N6 has been isolated from domestic poultry in the year 201015. The environmental sample from which the present virus was isolated was from the vicinity of a water body where mixed flocks of migratory water birds were present. It has been reported that the rates of isolation of AI viruses from environmental samples are low12132425. The isolated virus was a novel reassortant and phylogenetically divergent than the earlier H4N6 viruses from India. AI H4N6 virus isolated from India in the year 2010 belongs to the Eurasian lineage13. Their PB2, PB1, PA, NP, NA and M genes showed high PNIs with H4N6-1722769, indicating similarity and/or common ancestry. However, the PNIs of HA for the previous isolates were low, and they formed a separate group as compared to H4N6-1722760 in the phylogenetic trees indicating divergence (Table III and Fig. 1A). For the NS genes, the PNIs of the two chicken isolates were low and they belonged to the allele A, also indicating divergence. Interestingly, the PNI of the NS gene from the duck isolate, A/Duck/India/10736/2009, showed high similarity with H4N6-1722760 and grouped within the same allele (allele B), this indicated common ancestory for the NS gene (Table III and Fig. 1A). Previous reports of H4N6 virus from across the globe indicate isolation from ducks such as Teals, Mallard and Muscovy, and even from pigs9. It has been shown that the pH of the faecal droppings may affect virus isolation and that acidic pH has detrimental impact on AI virus isolation16. The pH of droppings shows variation among different bird species based on diet. It has been experimentally shown that acidic pH hampers virus isolation. The pH of the dropping from which the virus was isolated was basic (pH 8.0) favouring survival of the virus in faecal dropping enabling virus isolation.
The close relatedness of H4N6-1722760 with viruses from Asia and Europe indicated the probable role of migratory birds in its transmission since India lies in the central Asian flyway, which facilitates intermixing of different migratory bird species. Notably, ringing and tracking studies conducted on Brown-headed gulls have shown that the birds migrate from their breeding grounds in China to overwinter in Thailand and Cambodia; with Bangladesh, India, Myanmar and Vietnam as stopover sites28. In addition, ring recovery studies report a direct migratory connection between India and Russia and between India and Europe29. During the present surveillance activity, migratory, resident, and also resident migratory bird species were observed at the study site. This assertively indicates intermingling of different types of bird species which might lead to the possible transmission of the virus, since it is believed that interactions between migratory birds and resident domestic poultry in close proximity could be a factor in the spread of AI viruses30.
It has been demonstrated experimentally that avian H4 virus can adapt to mammals by point mutations in PB2 or HA10. Furthermore, the seroprevalence of antibodies against AI H4 viruses in poultry workers highlights a potential threat to public health11. In this regard, there is a dearth of extensive surveillance studies on migratory birds and serosurveillance studies among at-risk populations from India. The limitation of the present study was that the sample size was small.
In conclusion, the LPAI H4N6 virus isolated from Maharashtra, India, was found to be a reassortant and of low pathogenicity. Similarity of the virus with other Eurasian viruses signifies the possible role of wild and migratory birds in the transmission and spread of LPAI viruses in India. Further, molecular deviation of the current H4N6 virus from the previously isolated viruses from poultry indicates the possibility of independent introduction. Since the possibility of human infections with H4 viruses cannot be ruled out, there is a need for continuous AI surveillance in wild and migratory birds along with poultry, in India to study and monitor continuing virus evolution at the animal-human interface.
Acknowledgment
Authors acknowledge Servshree AA Deshpande & RG Bangale for help in the laboratory work; Shree JPN Babu for the support on field; Director, ICMR-National Institute of Virology for constructive suggestions. Authors also acknowledge WHO Collaborating Center for Reference and Research on Influenza, Melbourne, Australia for providing reference virus strains for the NAI assay.
Financial support & sponsorship: This work was supported by ICMR-National Institute of Virology, Pune, Maharashtra and Department of Health Research, Ministry of Health and Family Welfare, Government of India. The funding agency does not have any role in study design, collection, analysis and interpretation of data, in the writing of the report or in the decision to submit the article for publication.
Conflicts of Interest: None.
References
- A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3-13.
- [Google Scholar]
- Prevalence of the C-terminal truncations of NS1 in avian influenza A viruses and effect on virulence and replication of a highly pathogenic H7N1 virus in chickens. Virulence. 2016;7:546-57.
- [Google Scholar]
- Emergence of a highly pathogenic avian influenza virus from a low-pathogenic progenitor. J Virol. 2014;88:4375-88.
- [Google Scholar]
- Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247-50.
- [Google Scholar]
- Isolation and characterization of H4N6 avian influenza viruses from mallard ducks in Beijing, China. PLoS One. 2017;12:e0184437.
- [Google Scholar]
- Avian influenza viruses detected in birds in Sub-Saharan Africa: A systematic review. Viruses. 2020;12:993.
- [Google Scholar]
- Active virological surveillance in backyard ducks in Bangladesh: Detection of avian influenza and gammacoronaviruses. Avian Pathol. 2020;49:361-8.
- [Google Scholar]
- Transition in genetic constellations of H3N8 and H4N6 low-pathogenic avian influenza viruses isolated from an overwintering site in Japan throughout different winter seasons. Arch Virol. 2020;165:643-59.
- [Google Scholar]
- Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology. 2017;511:56-65.
- [Google Scholar]
- Mutations in PB2 and HA enhanced pathogenicity of H4N6 avian influenza virus in mice. J Gen Virol. 2020;101:910-20.
- [Google Scholar]
- Evidence of infection with H4 and H11 avian influenza viruses among Lebanese chicken growers. PLoS One. 2011;6:e26818.
- [Google Scholar]
- Genetics, receptor binding, replication, and mammalian transmission of H4 avian influenza viruses isolated from live poultry markets in China. J Virol. 2016;90:1455-69.
- [Google Scholar]
- Characterization of the influenza A H5N1 viruses of the 2008-09 outbreaks in India reveals a third introduction and possible endemicity. PLoS One. 2009;4:e7846.
- [Google Scholar]
- An avian influenza A(H11N1) virus from a wild aquatic bird revealing a unique Eurasian-American genetic reassortment. Virus Genes. 2010;41:14-22.
- [Google Scholar]
- Avian influenza surveillance reveals presence of low pathogenic avian influenza viruses in poultry during 2009-2011 in the West Bengal State, India. Virol J. 2012;9:151.
- [Google Scholar]
- Morphological and biochemical characteristics of avian faecal droppings and their impact on survival of avian influenza virus. Food Environ Virol. 2018;10:99-106.
- [Google Scholar]
- WHO manual on animal influenza diagnosis and surveillance. Geneva: WHO; 2002.
- Use of reverse transcriptase PCR to subtype N1 to N9 neuraminidase genes of avian influenza viruses. J Clin Microbiol. 2009;47:2301-3.
- [Google Scholar]
- Influenza virus titration, antigenic characterization, and serological methods for antibody detection. In: Kawaoka Y, Neumann G, eds. Influenza virus methods in molecular biology: Methods and protocols. New York, USA: Humana Press; 2012. p. :25-51.
- [Google Scholar]
- Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virol J. 2012;9:251.
- [Google Scholar]
- Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 2004;62:37-45.
- [Google Scholar]
- GISAID: Global initiative on sharing all influenza data – From vision to reality. Euro Surveill. 2017;22:30494.
- [Google Scholar]
- Characterizations of H4 avian influenza viruses isolated from ducks in live poultry markets and farm in Shanghai. Sci Rep. 2016;6:37843.
- [Google Scholar]
- A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J Virol. 2011;85:652-62.
- [Google Scholar]
- The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol. 2006;80:11115-23.
- [Google Scholar]
- A new influenza virus virulence determinant: The NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381-6.
- [Google Scholar]
- Role of the B allele of influenza A virus segment 8 in setting mammalian host range and pathogenicity. J Virol. 2016;90:9263-84.
- [Google Scholar]
- Satellite tracking on the flyways of brown-headed gulls and their potential role in the spread of highly pathogenic avian influenza H5N1 virus. PLoS One. 2012;7:e49939.
- [Google Scholar]
- Wild bird surveillance around outbreaks of highly pathogenic avian influenza A(H5N8) virus in the Netherlands, 2014, within the context of global flyways. Euro Surveill. 2015;20:21069.
- [Google Scholar]
- Test-modeling highly pathogenic avian influenza transmission in wild birds and poultry in West Bengal, India. Sci Rep. 2013;3:1-8.
- [Google Scholar]






