Translate this page into:
Perspectives for repurposing drugs for the coronavirus disease 2019
For correspondence: Dr Sarah S. Cherian, Department of Bioinformatics, ICMR-National Institute of Virology, 20-A,Dr Ambedkar Road, Pune 411 001, Maharashtra, India e-mail: cheriansarah@yahoo.co.in
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The newly emerged 2019 novel coronavirus (CoV), named as severe acute respiratory syndrome CoV-2 (SARS-CoV-2), like SARS-CoV (now, SARS-CoV-1) and Middle East respiratory syndrome CoV (MERS-CoV), has been associated with high infection rates with over 36,405 deaths. In the absence of approved marketed drugs against coronaviruses, the treatment and management of this novel CoV disease (COVID-19) worldwide is a challenge. Drug repurposing that has emerged as an effective drug discovery approach from earlier approved drugs could reduce the time and cost compared to de novo drug discovery. Direct virus-targeted antiviral agents target specific nucleic acid or proteins of the virus while host-based antivirals target either the host innate immune responses or the cellular machineries that are crucial for viral infection. Both the approaches necessarily interfere with viral pathogenesis. Here we summarize the present status of both virus-based and host-based drug repurposing perspectives for coronaviruses in general and the SARS-CoV-2 in particular.
Keywords
Coronavirus
COVID-19
drugs
host-based
repurposing
severe acute respiratory syndrome coronavirus 2
virus-based
Introduction
Coronaviruses (CoVs) belong to the family Coronaviridae and are enveloped, single-stranded, positive-sense RNA viruses1. The CoVs are seen to be distributed in mammals as well as in humans causing mild infections. However, the severe acute respiratory syndrome CoV (SARS-CoV) and the Middle East respiratory syndrome CoV (MERS-CoV) from zoonotic sources in 2002 and 2012, respectively, were responsible for high infection and mortality rates2. A novel CoV named as SARS-CoV-2, causative agent of the CoV disease 2019 (COVID-19), has caused 750,890 confirmed cases globally with 36,405 reported mortalities3. The SARS-CoV-2 belongs to the beta CoV genus which also includes the SARS-CoV-1 and the MERS-CoV. The lack of approved effective drug therapeutic protocols for CoVs would be a challenge for the treatment of the newly emerged COVID-19 infections worldwide.
Drug repurposing, which is defined as identifying alternative uses for approved or investigational drugs outside their defined indication, could be a possible way to overcome the time limitation of research and development needed to design a therapeutic drug to combat the pathogen4. Apart from having a lower risk of failure, most repurposed drugs have cleared phase I trials and require lower investment, but above all, the drug repurposing strategy drastically reduces the time frame for development5. The drug repurposing or repositioning approach thus can facilitate prompt clinical decisions at lower costs than de novo drug development. Though drug repurposing is sometimes based on chance observations, target-based repurposing of drugs depends on prior understanding of the precise molecular or cellular element that is recognized by the proposed drug67. The target may or may not essentially have the same mechanism of action in both the diseased states. Antivirals that can target the viral proteins or the key events in the viral life cycle, including virus-host cell interactions, replication, assembly and egress, would belong to this class. Drug repurposing to identify candidate drug compounds centred on the target-based criteria can thus be generally distinguished into virus- and host-based therapeutics. This review outlines the present status of both virus-based and host-based drug repurposing evaluations against the CoVs. The focus would be on the Food and Drug Administration (FDA)-approved marketed drugs or those under clinical trials against the CoVs in general, and the SARS-CoV-2 in particular.
Virus-based drug repurposing for coronaviruses
Virus-based antiviral agents target specific proteins of the virus. The major open reading frame, ORF1ab, of the SARS-CoV genome encodes the large replicase polyprotein pp1ab which forms the non-structural proteins, nsp1-16, while the structural proteins include S, E, M and N8910. The viral replication is facilitated by a replicase complex that involves processing of pp1ab by two cysteine proteases, namely the main protease (Mpro) or the 3C-like protease (3CLpro) and the secondary papain-like protease 2 (PL2pro)1112 (Figs 1 and 2). Mpro cleaves at 11 sites in the central and C-terminal regions, while PL2pro cleaves at three sites in the N-terminal regions of the polyprotein. Majority of the proteins and enzymes of CoVs vital for the replication process are potential drug targets.
![Schematic representation of the genomic organization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in comparison with bat-CoV RaTG 13, SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV). Below are the modelled three-dimensional structures of the major virus based antiviral targets [3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp) and papain-like protease (PL2pro)] based on SARS-CoV-1 templates obtained from Protein Data Bank. Also depicted is structure of the spike glycoprotein of SARS-CoV-2 released recently (6VSB.pdb). Per cent identity between coding regions of the specific viral genomes depicted was calculated using p-distance method of MEGA software v7.0 (https://www.megasoftware.net/). Source: Refs 913.](/content/175/2020/151/2-3/img/IJMR-151-160-g001.png)
- Schematic representation of the genomic organization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in comparison with bat-CoV RaTG 13, SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV). Below are the modelled three-dimensional structures of the major virus based antiviral targets [3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp) and papain-like protease (PL2pro)] based on SARS-CoV-1 templates obtained from Protein Data Bank. Also depicted is structure of the spike glycoprotein of SARS-CoV-2 released recently (6VSB.pdb). Per cent identity between coding regions of the specific viral genomes depicted was calculated using p-distance method of MEGA software v7.0 (https://www.megasoftware.net/). Source: Refs 913.
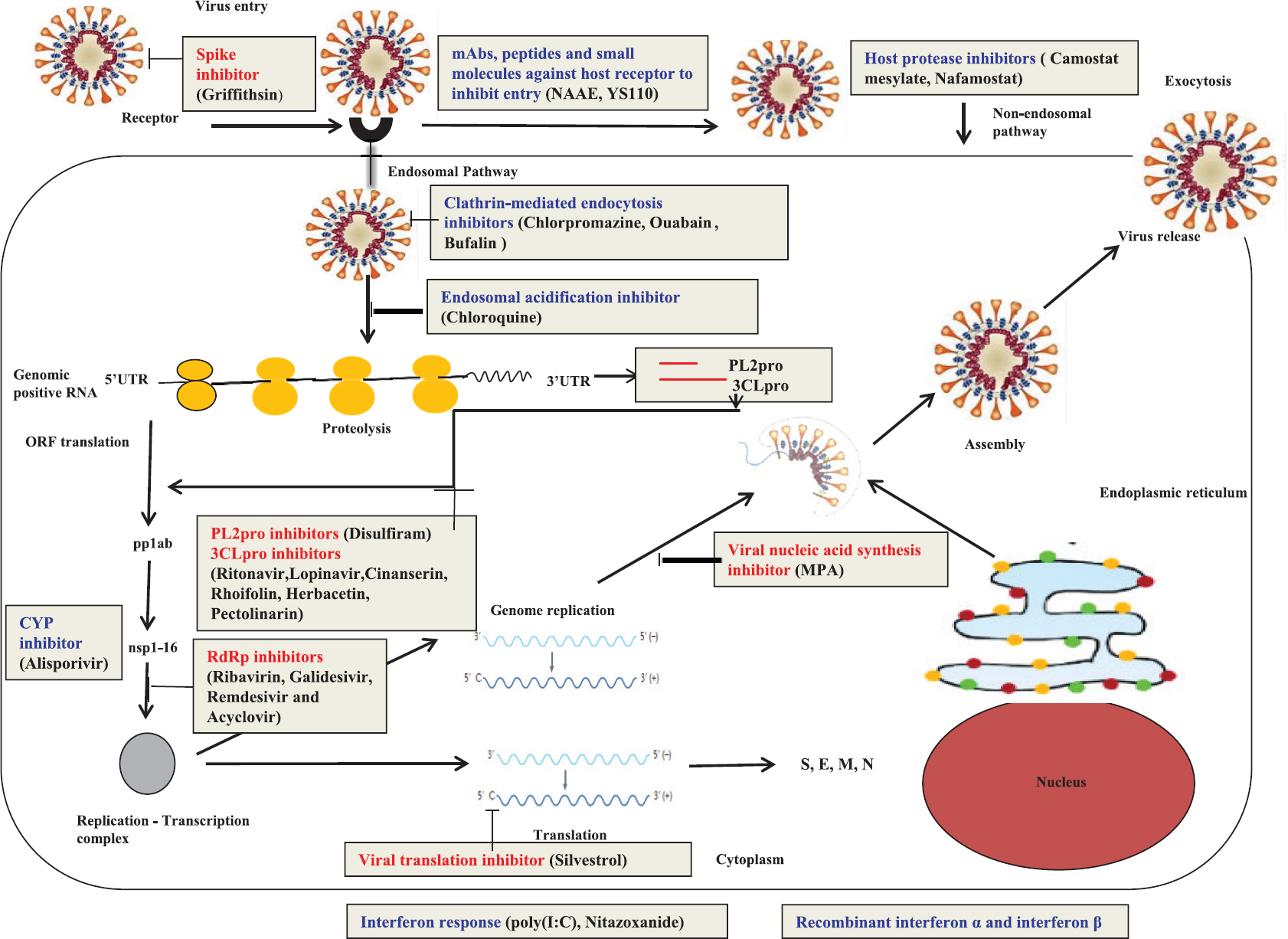
- Schematic representation of the coronavirus replication cycle depicting the potential therapeutics against different virus-based (red) and host-based (blue) targets for coronavirus drug repurposing. The drugs effective against the various targets are mentioned in the brackets. 3CLpro, cysteine-like protease; PL2pro, papain-like protease; nsp, non-structural protein; RdRp, RNA-dependent RNA polymerase; pp1ab, polyprotein ab; M, membrane protein; E, envelope protein; S, spike protein; N, nucleocapsid protein; UTR, untranslated region; ORF, open reading frame; MPA, mycophenolic acid; ERK-MAPK, extracellular signal-regulated kinase mitogen-activated protein kinase; poly(I:C), polyinosinic: polycytidylic acid; NAAE, N-(2-aminoethyl)-1-aziridine-ethanamine; YS110, a recombinant humanized IgG1 anti-DPP4 mAb; DPP4, dipeptidyl peptidase 4; CYP, cyclophilin. Source: Refs 8101415161718192021.
Main protease (Mpro)/ 3CLpro inhibitors - Lopinavir and/or lopinavir-ritonavir, cinanserin, herbacetin, rhoifolin and pectolinarin
The Mpro is a promising viral target for the design of drugs against SARS/MERS, as the polyprotein cleavage by the Mpro facilitates the formation of the RNA-dependent RNA polymerase (RdRp) and the helicase which are the major proteins of viral replication811122223. Various classes of protease inhibitors, such as halomethylketones, phthalhydrazide ketones, α, β-epoxyketones, glutamic acid and glutamine peptides with a trifluoromethylketone group, zinc or mercury conjugates, C2-symmetric diols, peptidomimetic-α, β-unsaturated esters, aldehydes, anilides, nitriles, pyrimidinone and pyrazole analogues, benzotriazole, N-phenyl-2-acetamide and biphenyl sulphone, are reported to inhibit the SARS-CoV-1 Mpro/ 3CLpro2425 (Fig. 2). Of these prospective Mpro inhibitors, the common FDA-approved ones are well-known HIV-1 protease inhibitors26. Among these, lopinavir and/or a ritonavir-boosted form of lopinavir has been reported to have anti-CoV activity in vitro and also has shown improved outcomes in non-human primates infected with MERS-CoV and in non-randomized trials with SARS patients27. Both lopinavir and ritanovir are under phase II/III clinical trials for MERS-CoV (NCT02845843)28. These are also reported to have activity against HCoV-229E, HCoV-NL63 and animal CoVs29.
Cinanserin (SQ 10,643) a serotonin antagonist, demonstrated antiviral activity against SARS-CoV-1, and the inhibition of replication was probably by blocking the activity of Mpro14. Flavonoids, herbacetin, rhoifolin and pectolinarin that are known to possess antioxidant effects associated with diseases such as cancer, Alzheimer's disease and atherosclerosis were also noted to efficiently inhibit SARS-CoV-1 Mpro15.
Papain-like protease (PLpro) inhibitor - Disulfiram
Disulfiram, which is an approved drug for the treatment of alcohol dependence, demonstrated in vitro inhibition of the PL2pro enzyme of SARS and MERS30. The study also provided future directions for the development of fragment-linked inhibitors for improving its potency31.
RNA-dependent RNA polymerase (RdRp) inhibitors - Ribavirin, immucillin-A/ galidesivir, remdesivir and acyclovir
The RdRp which is critical for CoV transcription and replication is involved in producing the genomic and subgenomic RNAs. Nucleoside analogues such as favipiravir, ribavirin, penciclovir, remdesivir and galidesivir are well-known RdRp inhibitors. A guanosine analogue, ribavirin, showed broad-spectrum antiviral activity against several viruses including respiratory syncytial virus, hepatitis C and E viruses (HCV, HEV), chikungunya and viral haemorrhagic fevers3233. Though the mechanism of action is not fully understood, it is hypothesized that the drug may be involved in the inhibition of mRNA capping or viral RNA synthesis. The in vitro antiviralactivityofribavirin was demonstrated against SARS-CoV-1 and MERS-CoV34 and in rhesus monkeys infected with MERS-CoV35. The drug has been used in the treatment of SARS and MERS patients, though the benefits are ambiguous. Further, in severely infected CoV patients, there could be side effects associated with high doses36.
Immucillin-A (galidesivir), an adenosine analogue, has been shown recently as a broad-spectrum RdRp inhibitor against several RNA viruses, such as paramyxoviruses, flaviviruses, togaviruses, bunyaviruses, arenaviruses, picornaviruses, filoviruses and also against SARS/MERS-CoVs37. Though it has been reported as a treatment option during the 2014-2016 West Africa Ebola virus epidemic, no data for animal/human were reported for CoVs until recently for the SARS-CoV-216.
Sheahan et al38 showed that another nucleoside analogue, remdesivir (GS-5734), presently under clinical trials for the Ebola virus, demonstrated inhibition of the replication of SARS-CoV-1 and MERS-CoV inprimary human airway epithelial cells. They also demonstrated broad-spectrum anti-CoV activity against bat-CoVs and human CoVs in primary human lung cells1738. In another recent study, remdesivir was shown to possess better in vitro antiviral efficacy against MERS-CoV in comparison to lopinavir and ritanovir1739. In mice, remdesivir improved pulmonary function with lower viral loads in the lungs both as a prophylactic and as a therapeutic1740.
Another nucleoside analogue, acyclovir that was modified by incorporating fleximers to increase its binding affinity has been reported to be effective in vitro against MERS-CoV and HCoV-NL633941, though to the best of our knowledge, no animal or human data are available.
Inhibitors of spike glycoprotein - Griffithsin
CoVs possess a surface structural spike glycoprotein (S) which is vital for interaction with the host cell receptor and subsequent virus entry into the cell. The S protein constitutes two subunits, the S1 (receptor-binding) and the S2 (membrane fusion) domains40. Griffithsin, a lectin extract red algae, has been reported to bind to oligosaccharides on the surface of various viral glycoproteins, including HIV glycoprotein 120 and SARS-CoV glycoproteins41.
Other inhibitors with unknown site of action - Resveratrol, amodiaquine, mefloquine, loperamide
Resveratrol, a natural compound from grape, which is in a clinical phase for heart and other diseases, was also reported to effectively inhibit MERS-CoV in vitro by downregulation of the apoptosis induced by the virus42. The possible site of action was suggested to be the nucleocapsid protein. Amodiaquine and mefloquine, antimalarial drugs, were also found to be effective against MERS-CoV43. Loperamide, an antidiarrhoeal agent that was identified by the screening of an FDA-approved compound library, showed in vitro antiviral activity against MERS44.
Inhibitors of viral nucleic acids - Mycophenolic acid
Viral nucleic acids are mainly composed of nucleosides and nucleotides. The drugs that target these have mycophenolic acid (MPA) as the active compound and inhibit inosine monophosphate dehydrogenase and guanine monophosphate synthesis45. Broad-spectrum activity has been reported by MPA against a broad range of viruses including orthohepadnaviruses (hepatitis B), flaviviruses (HCV), arboviruses and CoVs. MPA possessed anti-MERS-CoV activity in vitro, though it was shown to result in a worsened outcome in the marmoset primate model26. Treatment of renal transplant recipients with MPA resulted in severe MERS46. Combination therapy with interferon beta-1b (IFN-β-1b) was, however, reported to be synergistic in vitro47, implying that monotherapy with the drug might not be useful for treating CoVs.
Host-based drug repurposing for coronaviruses
Specific host factors are utilized by CoVs for entry and replication. The anti-CoV potential of monoclonal antibodies (mAbs) evoked against the receptor binding domain (RBD) of S1 subunit and fusion inhibitors which target the S2 subunit has been reported in in vitro and/or in vivo studies484950. SARS-CoVs and HCoV-NL63 preferably utilize the angiotensin-converting enzyme 2 (ACE2) host receptor while dipeptidyl peptidase 4 (DPP4) is used by MERS-CoV5152 for entry. The further entry of CoVs into host cells includes the cell surface and/or endosomal pathways which are via host proteases such as transmembrane protease serine 2 (TMPRSS2) that cleave and activate viral S protein53. Inhibitors of these host proteases can prevent this proteolytic cleavage, partially blocking cell entry. Further, a group of drugs can target the endocytosis or cell entry44 (Fig. 2).
The innate IFN response of the host also has therapeutic potential as it controls viral replication after infection1854. Additional pathways of cell signalling have also been noted as possible therapeutic targets for CoVs55. These classes of inhibitors are discussed below.
Inhibitors targeting endocytosis or cell entry - Chlorpromazine, ouabain, bufalin, chloroquine
Chlorpromazine, an antipsychotic/tranquilizer drug, is also known to affect the assembly of clathrin-coated pits at the plasma membrane44. It showed broad-spectrum in vitro activity against viruses such as HCV, alphaviruses, SARS-CoV-1 and MERS-CoV. Ouabain and bufalin, examples of a class of steroids which bind sodium- or potassium-transporting ATPase subunit α1, also inhibited the endocytosis of MERS-CoV mediated by clathrin56. However, very high EC50/Cmax (half-maximal effective concentration value/peak serum concentration level) ratios at the typical dosages or toxicity, limit the clinical use of these endocytosis inhibitors. Acidification of the endosome can also affect endocytosis. Chloroquine, an antimalarial drug, can increase the intracellular pH by directing protons into the lysosomes57. It possesses broad-spectrum in vitro antiviral activities against flaviviruses, HIV, Ebola, Nipah and numerous CoVs58. However, it did not show activity in SARS-CoV-infected mice59. The anti-CoV activity of different endocytosis inhibitors thus need further in vivo evaluation.
Inhibitors of host receptor mediated viral entry - N-(2-aminoethyl)-1-aziridine-ethanamine (NAAE), peptides, mAb YS110
Specific peptide inhibitors and monoclonal or polyclonal antibodies can be used to target the host receptor48. N-(2-aminoethyl)-1-aziridine-ethanamine, a small-molecule inhibitor and synthetic ACE2-derived peptides showed inhibition of ACE2 activity and cell fusion via the S protein of SARS-CoV-1 in vitro6061. However, these inhibitors have not been tested in CoV patients. Monoclonal antibodies (mAbs) such as anti-dipeptidyl peptidase 4 (DPP-4) have also been reported to block cell entry of MERS-CoV in vitro62. YS110, an anti-DPP4 recombinant humanized IgG1 mAb, used in a phase I clinical trial, was found to be well tolerated in patients with advanced malignancies19. However, considering that host cell receptor usage differs in different CoVs, the anti-CoV activity of these agents may be narrow-spectrum. Further, based on the vital biological functions of these receptors, the risks of immunopathology such as blood pressure regulation, glucose metabolism etc., would need assessment52.
Inhibitors of host proteases used for viral entry - Camostat mesylate, nafamostat
Camostat mesylate, a synthetic serine protease inhibitor, that is used to treat patients with chronic pancreatitis, works against the serine protease TMPRSS26364. It has shown broad-spectrum activity against enveloped RNA viruses such as CoVs and paramyxoviruses. Camostat mesylate is reported to inhibit SARS and MERS in ex vivo studies and improves the survival of mice infected with SARS6465. Nafamostat, another serine protease inhibitor used to treat disseminated intravascular coagulation and pancreatitis, blocked MERS-CoV infection by inhibiting TMPRSS2 in human airway epithelial Calu-3 cells6566.
Enhancers of host innate immune response - Interferons, polyinosinic: polycytidylic acid [poly(I:C)] and nitazoxanide
Though on viral infection suppression of the IFN response is an integral part for immune evasion, several viruses and CoVs are noted to be susceptible to IFN treatment. The effectiveness of recombinant IFN-β over IFN-α has been demonstrated by in vitro studies against both SARS and MERS67.IFN-α mediated reduction of viral titres was observed in SARS-CoV-infected in vivo models3559, while IFN-β administration via different routes was found to be effective in MERS-CoV in vivo models26. Combinations of IFN-α/β, ribavirin and lopinavir/ritonavir-boosted lopinavir for treatment of SARS/MERS patients, demonstrated varying benefits333668.
Another type I IFN enhancer, polyinosinic: polycytidylic acid [poly(I:C)], a dsRNA synthetic analogue, demonstrated reduction in viral load in MERS-CoV-infected BALB/c mice69. In phase II clinical trials, poly (I:C) was shown to be beneficial for patients suffering from malignant gliomas70.
Nitazoxanide, a synthetic derivative of nitrothiazolyl-salicylamide which is used as a treatment for parasitic infections, is an effective type I IFN inducer71. It has been shown to exhibit antiviral activities against several viral families and canine CoVs. Nitazoxanide was found to be safe in phase II and III clinical trials against HCV and influenza72.
Inhibitors of signaling pathways involved in viral replication - Cyclosporine, trametinib and others
Drugs interfering with the viral replication signaling pathways are noted to have broad spectrum activity against several viruses such as HCV, HIV, vesicular stomatitis virus, human papilloma virus, vaccinia virus and CoVs55. Cyclosporine, a calcineurin pathway inhibitor, inhibited a broad range of CoVs in vitro by interacting with the nsp1 protein and modulating immune response mediated by T cells73. The clinical application of this drug is, however, restricted due to immune-suppressive effects and a higher EC50/Cmax ratio at standard dose levels. Other calcineurin inhibitors such as alisporivir, have demonstrated activity against HCoV-NL6320.
The extracellular signal-regulated kinase (ERK) pathway mediates intracellular signals from membrane-associated Ras to the cytoplasmic kinase cascade Raf, Mek and Erk74. The kinase signaling pathway inhibitors, such as trametinib (Mek inhibitor), selumetinib (Erk inhibitor), everolimus, rapamycin, dasatinib and imatinib have also demonstrated anti-CoV effects through inhibition of early viral entry or post-entry events75. However, their toxicities may be a concern in severe infections.
Targeting viral translation - Silvestrol
Initiation of translation in many viruses happens through the usage of the host eukaryotic initiation factors (eIFs)76. The helicase eIF4A unwinds 5′-untranslated region of the mRNA, facilitating assembly of the translation pre-initiation complexes. A natural compound, silvestrol, being an inhibitor of eIF4A and reported to show anti-cancer activity77, demonstrated inhibition of MERS-CoV and HCoV-229E translation and replication in MRC-5 lung fibroblast cells78.
Current perspectives for COVID-2019
Comparison of the coding regions of SARS-CoV-2 showed that it possessed a similar genomic organization when compared to bat-SL-CoVZC45 and SARS-CoV-19 (Fig. 2). Sequence analysis further revealed good sequence identity with the bat and human CoVs in the different coding regions. Except for the spike glycoprotein of SARS-CoV-2 that differs from the other CoVs including SARS-CoV-1 spike protein1379, the catalytic pockets in the major non-structural viral enzymes are conserved at both the sequence and protein structural level across CoVs. Hence, repurposing of the promising MERS and SARS inhibitors for SARS-CoV-2 is a practical strategy16.
In vitro evaluations to test the antiviral potency of marketed drugs ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine and broad-spectrum RdRp inhibitors, remdesivir (GS-5734) and favipiravir (T-705) against SARS-CoV-2 were recently undertaken80. The findings have shown that remdesivir and chloroquine are more efficacious in comparison to the others. A patient from USA with COVID 2019 who was treated with remdesivir intravenously was reported to have recovered81. Phase III trials (NCT04252664, NCT04257656) of intravenous remdesivir are currently ongoing to assess the efficacy in patients with SARS-CoV-2. Chloroquine is under an open-label trial for SARS-CoV-2 (ChiCTR2000029609). In addition, randomized clinical trials have been initiated for SARS-CoV-2 with favipiravir (ChiCTRChiCTR2000029544, ChiCTR2000029600) and ribavirin in combination with pegylated IFN (ChiCTR2000029387).
Results following rapid sequencing of the SARS-CoV-2, combined with molecular modelling based on homologous templates82 have identified certain compounds along with lopinavir and ritonavir that may be efficacious. Phase III clinical trials have also been initiated to test the HIV protease inhibitors including lopinavir (NCT04252274, NCT04251871, NCT04255017, ChiCTR2000029539), ritonavir (NCT04251871, NCT04255017, NCT04261270), darunavir and cobicistat (NCT04252274) in patients infected with SARS-CoV-221. Another HIV protease inhibitor, ASC09F, in combination with oseltamivir is also in phase III clinical trial for SARS-CoV-2 (NCT04261270).
Arbidol (Umifenovir), a wide-spectrum antiviral drug inhibiting several flaviviruses and influenza viruses, whose mechanism of action is based on blocking crucial steps in virus- host cell interactions83, is under phase IV clinical trial for SARS-CoV-2 (NCT04260594, NCT04254874, NCT04255017). Oseltamivir, an influenza neuraminidase inhibitor84 is also under phase IV trial for SARS-CoV-2 (NCT04255017).
In the direction of host-based treatment strategies, randomized trials are underway for SARS-CoV-2 using recombinant IFNs (NCT04251871, ChiCTR2000029638)19. In another study, an artificial intelligence-based knowledge graph comprising systematically curated medical data, was searched for approved drugs against SARS-CoV-285. Baricitinib, a janus kinase inhibitor, that was consequently identified, is a high-affinity AP2-associated protein kinase 1-binding drug which also interacts with a kinase regulator of endocytosis. Baricitinib has thus been suggested as a potential treatment for COVID-19 disease as it has the ability to reduce viral infection in lung cells.
Molecular docking studies undertaken
We analyzed the binding potential of HIV-1 protease inhibitors, lopinavir and ritanovir against the 3CLpro of SARS-CoV-2, using computational docking studies. This would help gain insight into the molecular mode of action of these drugs which are under clinical trials against the SARS-CoV-2 and also estimate the comparative inhibitory potency of the FDA-approved HIV protease inhibitors to the SARS-CoV-2.
The Mpro of CoVs cleaves substrates by recognizing the sequence motif (small)-X-(L/F/M)-Q↓(G/A/S)-X (X → any amino acid; ↓ cleavage site) and specifically the P1 site of the substrate requires a Gln (Q)8687. The X-ray structure of SARS-CoV-1 3CLPro dimer bound with aza peptide epoxide (APE) as an inhibitor, (2A5K.pdb) was used for the modelling studies. The peptide showed major specificity to the S2 subsite and partial specificity to the S4 subsite of 3CLpro12. We detached the APE from the crystal structure complex and re-docked it computationally using the same protocol as for the two selected study inhibitors to obtain the docking score and it was found to be −8.27 Kcal/mol. The two inhibitors in this study had better binding potential (Fig. 3) when compared to APE. Comparison of the docked poses reveals that lopinavir occupies the S1' and S1 subsites with excellent complementarity while ritanovir occupies the S3 and S4 subsites with excellent complementarity through the benzene and 2' isopropyl thiozole groups respectively. These structural features indicate the possible mechanism by which these inhibitors can block the function of the SARS-CoV-2 3CLpro. The peptide substrate cleavage sites for SARS-CoV 3CLpro are noted to be at P1↓ P1' and P3↓P48889, the occupancy at the respective active site cavities would be crucial for competitive inhibition of the polyprotein substrate. Based on this requirement, the findings are suggestive that ritonavir and lopinavir may have good potential for repurposing as SARS-CoV-2 protease inhibitors. Molecular dynamics simulation studies for the complexes obtained in this study would be essential to identify specific interactions between the enzyme and drug in the stable complexes and observe the hydrogen bond pattern, especially in the presence of solvent molecules. Additionally, studies need to be undertaken for the binding analyses of the other protease inhibitors, specific RdRp inhibitors and inhibitors of other enzymatic targets. The results would help gain an in-depth understanding of the relative binding affinity and design of derivatives with greater binding potential at the enzyme active site.
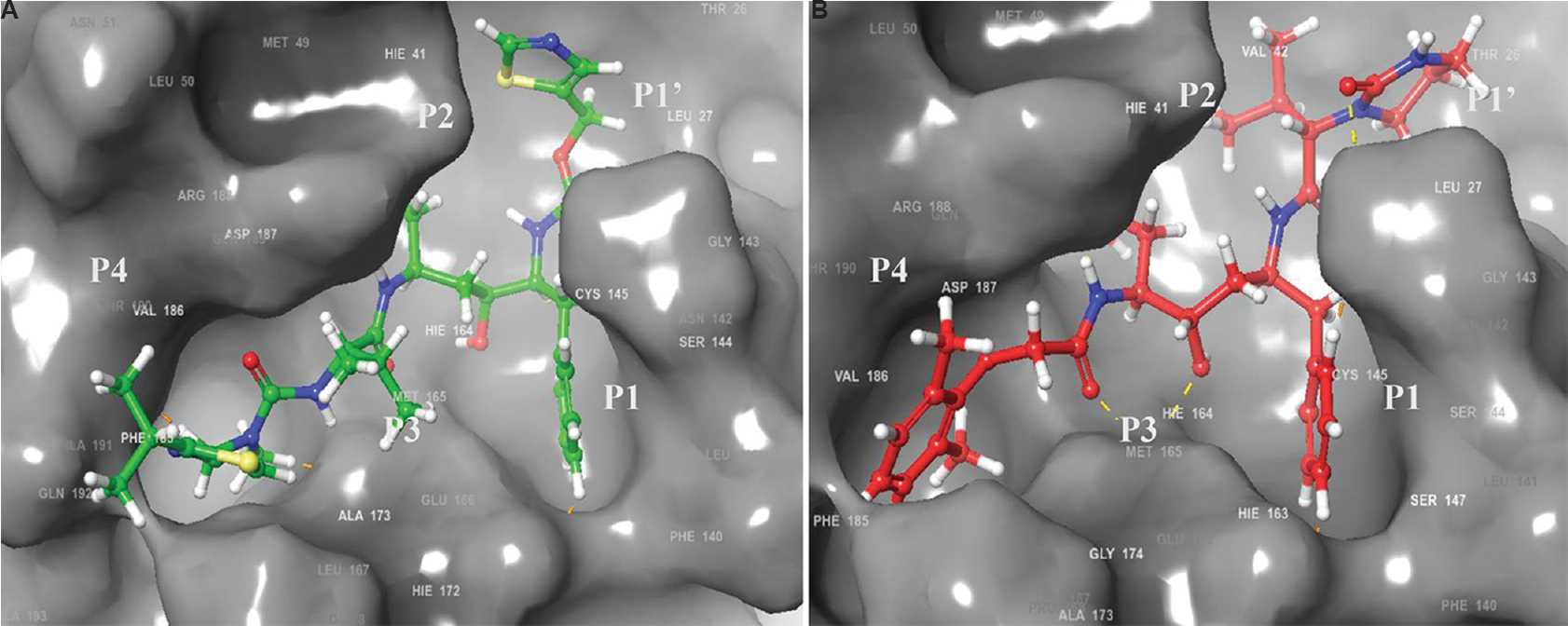
- Docking interaction analysis of HIV inhibitors in the substrate binding cavity of modelled severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (3C-like protease). (A) ritonavir (Docking score = −11.29 kcal/mol); (B) lopinavir (Docking score = −9.6 kcal/mol). The P4–P1' side chains of the inhibitor are labelled. Comparison of the docked poses of the inhibitors reveals that the occupancy at the respective active site cavity subsites corresponding to P1' and P1 for lopinavir and P3 and P4 for ritanovir that are crucial for competitive inhibition of the polyprotein substrate, are good.
Conclusions
This review presented the information with respect to repurposing of FDA-approved drugs as well as those under clinical trials for SARS-CoV-1 and MERS-CoVs, wherein a lot of effort had gone in during the last decade or more. This knowledge has in fact, formed the basis for efforts towards drug repurposing for the SARS-CoV-2 as well. As highlighted in this review, phase III clinical trials of a few drugs have been initiated, though most of these are notably targeting the virus directly, essentially the RdRp or the chymotrypsin-like protease 3CLpro. The spike glycoprotein also needs be explored as a target for the SARS-CoV-2 as the S1 domain of this virus deviates from the other human CoVs. It is thus important that the spike protein should be considered as a potential SARS-CoV-2 therapeutic target. On the other hand, considering that the strategy of targeting viral proteins is vulnerable to the emergence of viral resistance, other coronavirus targets such as the papain-like protease, helicase etc., also need to be attempted for drug repurposing. Further, several more of the potential SARS and/or MERS host-based inhibitors should be assessed against SARS-CoV-2. The ongoing vigorous efforts would help develop broad-spectrum anti-CoV agents against SARS-CoV-2.
Acknowledgment
The second author (MA) acknowledges the Indian Council of Medical Research (ICMR), New Delhi, for Senior Research Fellowship (SRF No. BIC/12(30)/2013). Technical assistance in the form of the schematic representations by Ms Bhagyashri Kasabe, Senior Research Fellow in the ICMR-funded project (Grant No. VIR/32/2019/ECD-1), and Shri Chandan Saini, Bioinformatics Group, ICMR-National Institute of Virology, Pune, is acknowledged.
Financial support & sponsorship: This work was funded by the ICMR-National Institute of Virology, Pune
Conflicts of Interest: None.
References
- Progress in anti-SARS coronavirus chemistry, biology and chemotherapy. Annu Rep Med Chem. 2007;41:183-96.
- [Google Scholar]
- Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52(Pt 8):715-20.
- [Google Scholar]
- Coronavirus disease 2019 (COVID-19) Situation Report - 71. Available from: https://wwwwhoint/docs/default-source/coronaviruse/situation-reports/20200331-sitrep-71-covid-19pdfsfvrsn=4360e92b _4
- Drug repurposing: Progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41-58.
- [Google Scholar]
- Drug repurposing approaches for the treatment of influenza viral infection: Reviving old drugs to fight against a long-lived enemy. Front Immunol. 2019;10:531.
- [Google Scholar]
- Drug repositioning: Identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673-83.
- [Google Scholar]
- Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465-522.
- [Google Scholar]
- Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565-74.
- [Google Scholar]
- Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327-47.
- [Google Scholar]
- Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol. 2001;75:6676-81.
- [Google Scholar]
- Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J Mol Biol. 2005;353:1137-51.
- [Google Scholar]
- Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-3.
- [Google Scholar]
- Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J Virol. 2005;79:7095-103.
- [Google Scholar]
- Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35:145-51.
- [Google Scholar]
- Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149-50.
- [Google Scholar]
- Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222.
- [Google Scholar]
- Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4:e00165-13.
- [Google Scholar]
- First-in-human phase 1 of YS110, a monoclonal antibody directed against CD26 in advanced CD26-expressing cancers. Br J Cancer. 2017;116:1126-34.
- [Google Scholar]
- Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184:44-53.
- [Google Scholar]
- Clinical trials on drug repositioning for COVID-19 treatment. Rev Panam Salud Publica. 2020;44:e40.
- [Google Scholar]
- Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305-15.
- [Google Scholar]
- Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr Pharm Des. 2006;12:4573-90.
- [Google Scholar]
- Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry. 2007;46:8744-52.
- [Google Scholar]
- Antiviral drugs specific for coronaviruses in preclinical development. Curr Opin Virol. 2014;8:45-53.
- [Google Scholar]
- Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J -Xianggang Yi Xue Za Zhi. 2003;9:399-406.
- [Google Scholar]
- Treatment With lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904-13.
- [Google Scholar]
- Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antivir Ther. 2016;21:455-9.
- [Google Scholar]
- Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 2018;150:155-63.
- [Google Scholar]
- Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorg Med Chem. 2019;27:1981-9.
- [Google Scholar]
- Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet. 2003;361:1615-7.
- [Google Scholar]
- Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: A retrospective cohort study. Lancet Infect Dis. 2014;14:1090-5.
- [Google Scholar]
- Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg Infect Dis. 2004;10:581-6.
- [Google Scholar]
- Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313-7.
- [Google Scholar]
- Clinical management and infection control of SARS: Lessons learned. Antiviral Res. 2013;100:407-19.
- [Google Scholar]
- Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402-5.
- [Google Scholar]
- Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017:9. pii: eaal3653
- [Google Scholar]
- Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity. Bioorg Med Chem Lett. 2015;25:2923-6.
- [Google Scholar]
- Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067.
- [Google Scholar]
- Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol. 2010;84:2511-21.
- [Google Scholar]
- Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis. 2017;17:144.
- [Google Scholar]
- Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014;58:4885-93.
- [Google Scholar]
- Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875-84.
- [Google Scholar]
- Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67:606-16.
- [Google Scholar]
- Distinct immune response in two MERS-CoV-infected patients: Can we go from bench to bedside? PLoS One. 2014;9:e88716.
- [Google Scholar]
- Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J Gen Virol. 2014;95:571-7.
- [Google Scholar]
- Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J Virol. 2013;87:13134-40.
- [Google Scholar]
- Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6:234ra59.
- [Google Scholar]
- Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2015;112:8738-43.
- [Google Scholar]
- Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450-4.
- [Google Scholar]
- Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251-4.
- [Google Scholar]
- Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87:12552-61.
- [Google Scholar]
- Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5:e01174-14.
- [Google Scholar]
- The SARS-coronavirus-host interactome: Identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331.
- [Google Scholar]
- ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J Virol. 2015;89:4434-48.
- [Google Scholar]
- Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69.
- [Google Scholar]
- In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun. 2004;323:264-8.
- [Google Scholar]
- Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir Chem Chemother. 2006;17:275-84.
- [Google Scholar]
- Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44:903-6.
- [Google Scholar]
- Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15-25.
- [Google Scholar]
- Inhibition of Middle East respiratory syndrome coronavirus infection by anti-CD26 monoclonal antibody. J Virol. 2013;87:13892-9.
- [Google Scholar]
- Efficacy of camostat mesilate against dyspepsia associated with non-alcoholic mild pancreatic disease. J Gastroenterol. 2010;45:335-41.
- [Google Scholar]
- Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76-84.
- [Google Scholar]
- Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60:6532-9.
- [Google Scholar]
- Use of a synthetic protease inhibitor for the treatment of L-asparaginase-induced acute pancreatitis complicated by disseminated intravascular coagulation. Ann Hematol. 1992;64:249-52.
- [Google Scholar]
- Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2013;3:1686.
- [Google Scholar]
- Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-94.
- [Google Scholar]
- Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111:4970-5.
- [Google Scholar]
- A multi-institution phase II study of poly-ICLC and radiotherapy with concurrent and adjuvant temozolomide in adults with newly diagnosed glioblastoma. Neuro Oncol. 2010;12:1071-7.
- [Google Scholar]
- Nitazoxanide: A first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94-103.
- [Google Scholar]
- Influenza antivirals currently in late-phase clinical trial. Influenza Other Respir Viruses. 2017;11:240-6.
- [Google Scholar]
- Suppression of coronavirus replication by cyclophilin inhibitors. Viruses. 2013;5:1250-60.
- [Google Scholar]
- Extracellular-regulated kinases: Signaling from ras to ERK substrates to control biological outcomes. Adv Cancer Res. 2018;138:99-142.
- [Google Scholar]
- Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59:1088-99.
- [Google Scholar]
- Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:7029-36.
- [Google Scholar]
- Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat Prod Rep. 2014;31:924-39.
- [Google Scholar]
- Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 2018;150:123-9.
- [Google Scholar]
- Peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein HR1 and HR2 domains. Int J Mol Sci 2018:19. pii: E487
- [Google Scholar]
- Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269-71.
- [Google Scholar]
- First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929-36.
- [Google Scholar]
- Coronavirus COVID-19 (Wuhan coronavirus and 2019-NCOV)- what we can find out on a structural bioinformatics level. Available from: https://innophorecom/2019-ncov/
- Arbidol (Umifenovir): A broad-spectrum antiviral drug that inhibits medically important arthropod-borne flaviviruses. Viruses 2018:10. pii: E184
- [Google Scholar]
- Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza Other Respir Viruses. 2013;7(Suppl 1):25-36.
- [Google Scholar]
- Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30-1.
- [Google Scholar]
- From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085-96.
- [Google Scholar]
- Conservation of substrate specificities among coronavirus main proteases. J Gen Virol. 2002;83:595-9.
- [Google Scholar]
- Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J Virol. 2012;86:11754-62.
- [Google Scholar]
- Recent developments on coronavirus main protease/3C like protease inhibitors. Recent Pat Antiinfect Drug Discov. 2013;8:150-6.
- [Google Scholar]






