Translate this page into:
Utility of a modified heat inactivation method for direct detection of SARS-CoV-2 by RT-qPCR in viral transport medium bypassing RNA extraction: A preliminary study
*For correspondence: lolekavita37@yahoo.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Sir,
The emergence of SARS-CoV-2 as a pandemic and the subsequent lockdowns to contain the spread of the infection has led to a situation wherein there is a supply deficit of critical reagents required for detection of the infection1. With regard to molecular diagnosis, extraction of the viral RNA is the first step and is usually performed using silica column-based methods or magnetic bead-based methods. Due to supply deficit, alternative RNA extraction methods are being explored. Investigation of direct application of heat inactivated clinical samples has gained momentum2. However, heat inactivation might cause coagulation of the proteins in the nasopharyngeal or oropharyngeal swab containing viral transport medium (VTM), leading to trapping of the viral RNA. Moreover, pipetting minute volume of the sample which becomes sticky due to heat inactivation is also difficult. In the present study, we explored the utility of protease enzyme and dithiothreitol (DTT) treatment prior to heat inactivation in releasing the viral RNA and its subsequent use in real-time reverse transcription-polymerase chain reaction (RT-qPCR) for detection of the SARS-CoV-2. DTT is a reducing agent which is known to break the disulphide bonds and affects the tertiary structures of the proteins. Thus, DTT can inactivate RNases and preserve the RNA molecules3, while protease cleaves the peptide bonds, thereby releasing amino acids and helping in liquefying the viscous sample4.
This study was conducted at the ICMR-National Institute of Virology (ICMR-NIV), Pune, India, during April-June 2020. The study protocol was approved by the Institutional Human Ethics Committee. For protease/DTT/heat inactivation (P/D/HI) treatment, 100 μl of the sample from VTM tube was taken and mixed with 10 μl (1 mg/ml) of protease (Qiagen, Germany) and 1 μl of 0.1 M DTT (Thermo Fisher Scientific, USA) and incubated at room temperature for 10 min, followed by 56°C for 15 min and then 95°C for 10 minutes. From the heat inactivated sample, 5 μl was subjected to the detection of SARS-CoV-2 using ICMR-NIV RT-qPCR assay using AgPath One-Step RT-PCR Kit (Life Technologies, USA) as described earlier5. Among the samples tested at the ICMR-NIV for SARS-CoV-2, 157 SARS-CoV-2-positive samples and 292 negative samples were subjected to P/D/HI treatment and used in RT-qPCR. Earlier, RNA was extracted from these samples using MagMAX-96 Viral RNA Isolation Kit (Thermo Fisher Scientific, USA) in an automated RNA extractor and was tested by ICMR-NIV RT-qPCR assay5. P/D/HI treatment resulted in the detection of SARS-CoV-2 in 64.3 per cent of the positive samples. Among the negative samples, only one sample showed false-positive result (Table). The presence of inhibitors in the samples might vary and could have resulted in more false-negative results.
| Heat inactivation method | Total samples tested* | Total positive samples tested* | Numbers detected as positive | Sensitivity (%) with 95% confidence interval | Total negative samples tested* | Numbers detected as negative | Specificity (%) with 95% confidence interval |
|---|---|---|---|---|---|---|---|
| Protease and DTT treatment followed by heat inactivation without dilution | 449 | 157 | 101 | 64.3 (56.6-71.4) | 292 | 291 | 99.7 (98.1-99.9) |
| Protease and DTT treatment followed by heat inactivation with dilution in the ratio of 1:1 | 324 | 211 | 196 | 92.9 (88.6-95.6) | 113 | 105 | 92.9 (86.6-96.4) |
*The samples were considered as positive or negative for SARS-CoV-2 by ICMR-NIV RT-qPCR assay performed on RNA extracted using MagMAX-96 Viral RNA Isolation Kit (Thermo Fisher Scientific, USA) in an automated RNA extractor. DTT, dithiothreitol
To decrease the concentration of PCR inhibitors and increase the sensitivity of detection of SARS-CoV-2, the P/D/HI-treated sample was diluted with an equal volume of nuclease-free water, and then 5 μl of the mixture was subjected to ICMR-NIV RT-qPCR assay using AgPath One-Step RT-qPCR Kit. This modified assay was used for the direct detection of SARS-CoV-2 without RNA extraction in 211 positive samples and 113 negative samples. With the modified method, the sensitivity increased to 92.9 per cent with a specificity of 92.9 per cent (Table).
Among these samples tested, 46 samples were tested by both the methods. The proportion of positive samples detected by different methods was compared by McNemar's test. Cycle threshold (Ct) values for the samples positive by both the methods were compared using paired t test. Statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA).
P/D/HI treatment without dilution detected only 16 samples (34.8%) as positive, whereas P/D/HI treatment with dilution led to the detection of 41 samples (89.1%) as positive (McNemar's P <0.001). When the Ct values were compared between both the methods for E, ORF1b, RdRp and RNase P genes, the Ct values were significantly lower in samples subjected to P/D/HI treatment with dilution as compared to that of undiluted P/D/HI-treated samples (Figure). The mean Ct values (with standard deviation) of the samples subjected to P/D/HI treatment with dilution but did not show amplification when subjected to P/D/HI treatment without dilution were 31.04±3.09, 31.12±2.49, 31.21±2.92 and 30.91±1.97 for E, ORF1b, RdRp and RNase P genes, respectively.
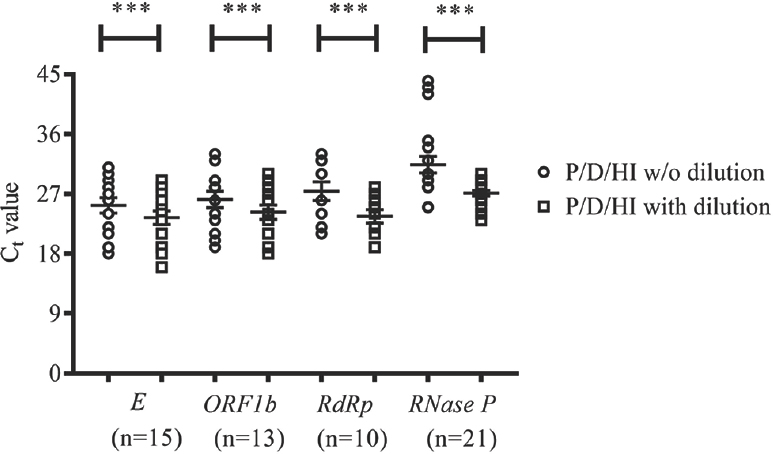
- Comparison of cycle threshold (Ct) values for different SARS-CoV-2 genes and internal control gene between samples subjected to protease/dithiothreitol/heat inactivation with and without dilution. The data are represented as aligned dot plot with lines for mean and standard error of the mean. For each gene, samples which showed Ct value in both treatments only were included for comparison. ***P<0.001 compared to without dilution.
The present results suggested that the modified heat inactivation protocol with the addition of protease and DTT followed by dilution might have resulted in reducing the presence of inhibitors for RT-qPCR. This modified method can be applied in the absence of RNA extraction kits. Although this method resulted in eight false-positive results, it is possible that these samples with RNA extraction might have yielded false-negative results due to the low viral load. However, it was not possible to cross-check the results by repeat testing with freshly extracted RNA due to the lack of sufficient quantity of clinical samples. Similar to this study, a study by Ladha et al6 has utilized an approach involving dilution of swab sample followed by addition of an extraction solution containing detergents and proteinase K and heat inactivation for direct RT-qPCR. In the present study, proteinase K was not used because it might not have got inactivated even at higher temperatures and could interfere with RT-qPCR.
Because all the P/D/HI-treated samples were tested using AgPath One-Step RT-PCR Kit, we tested whether P/D/HI-treated samples were compatible with other one-step RT-qPCR enzyme systems. P/D/HI-treated samples did not work with SuperScript™ III Platinum One-Step qRT-PCR Kit (Thermo Fisher Scientific, USA), whereas the samples worked with One-Step Primescript III RT-qPCR mix and One-Step PrimeScript RT-PCR Kit (Takara Bio, Japan), iTaq™ Universal Probes One-Step Kit (Bio-Rad, USA), TaqPath One-Step Multiplex Master Mix and TaqMan™ Fast Virus One-Step Master Mix (Thermo Fisher Scientific, USA). Moreover, the samples also worked with LabGun COVID-19 RT-PCR Kit (LabGenomics, Republic of Korea). The enzyme systems which have worked with P/D/HI-treated samples might be more robust and have increased tolerance to inhibitors. We also checked whether the P/D/HI treatment worked with samples collected in lysis buffers or those in molecular transport medium (MTM). However, the treatment did not yield positive results. It was possible that components such as chaotropic salt and detergent in the lysis buffer might have affected the enzyme activity and hence the results. A study has reported that samples collected in Hanks medium or saline water require RNA extraction7.
The utility of other proteases (Sigma-Aldrich, USA) or Pronase (Roche Diagnostics, Germany) was also tested. The treatment results were not affected by changing the proteases. However, the treatment conditions needed modifications according to the protease used. The method has its own limitations such as need for a more robust and tolerable enzyme system and collection of samples in VTM rather than in lysis buffers or MTM. The present study was a single-centre study and was done in an apex laboratory. However, further trials need to be conducted in multiple laboratory settings.
To conclude, the present study suggests that in the absence of RNA extraction kits, a modified heat inactivation protocol involving protease and DTT treatment followed by dilution of the sample may be useful if the samples are collected exclusively in VTM tubes.
Financial support & sponsorship: The study was funded by the Indian Council of Medical Research, Department of Health Research, Ministry of Health and Family Welfare, Government of India, New Delhi.
Conflicts of Interest: None.
References
- Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771-83.
- [Google Scholar]
- Endogenous RNase inhibitor contributes to stability of RNA in crude cell lysates: Applicability to RT-qPCR. Anal Biochem. 2016;513:21-7.
- [Google Scholar]
- An optimised protocol for isolation of RNA from small sections of laser-capture microdissected FFPE tissue amenable for next-generation sequencing. BMC Mol Biol. 2017;18:22.
- [Google Scholar]
- Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151:251-4.
- [Google Scholar]
- A 5-min RNA preparation method for COVID-19 detection with RT-qPCR medRxiv. 2020 doiorg/101101/2020050720055947
- [Google Scholar]
- SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J Clin Virol. 2020;128:104423.
- [Google Scholar]





