Translate this page into:
Identification of hepatitis B virus genotypes detected in Lahaul & Spiti district of Himachal Pradesh, India
For correspondence: Dr Pradip V. Barde, ICMR-National Institute of Virology, Central Zone, ICMR-NIRTH Campus, Nagpur Road, Garha, Jabalpur, Madhya Pradesh 482 003, India e-mail: pradip_barde@hotmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Infection by hepatitis B virus (HBV) results in acute or chronic hepatitis. Based on sequence differences of eight per cent or more, HBV is divided into 10 genotypes (A to J) and 35 sub-genotypes. Molecular characterization of the circulating HBV genome has helped in understanding the epidemiology and its clinical importance. Spiti valley in Himachal Pradesh, which shares its border with Tibet, is one of the most HBV prevalent areas in India. Since information about the circulating genotype/s of HBV in this area is limited, this study was conducted to identify the circulating HBV genotypes.
Methods:
The surface and partial reverse transcriptase gene regions were sequenced using 14 hepatitis B surface antigen-positive samples.
Results:
Out of the 14 hepatitis B surface antigen-positive samples 11 sample gave quality sequence for further analysis. All the 11 samples belonged to subtype ayw2. The phylogenetic and recombination analysis revealed that five out of 11 samples were of genotype CD1 and the rest six were of genotype D3.
Interpretation & conclusions:
The CD1 recombinant sub-genotype might have immigrated during past or present transcontinental migration between the adjacent countries. Further studies using full-genome sequencing and high sample size will be helpful to understand this epidemiology and to combat the high prevalence of HBV in the area.
Keywords
Genotype CD1
genotype/s
hepatitis B virus
India
recombinant genotype
Hepatitis B virus (HBV) infection of the liver is a serious health hazard affecting approximately 257 million people every year globally1. HBV can cause both acute and chronic liver diseases leading to high morbidity and mortality1. In India, the prevalence of HBV infection ranges between 4-7 per cent and an estimated 40 million people belonging to different geographies, communities to social and professional groups are reportedly HBV carriers, placing India in the intermediate HBV endemicity group2.
HBV belongs to the Hepadnaviridae family and contains a partial double-stranded (ds) circular DNA of approximately 3.2 kb. The compact circular genome encodes four overlapping genes, such as surface (S), core (C), regulatory (X) and polymerase (P) genes for regulation, replication and the formation of the nucleocapsid and the viral envelope3. The genetic heterogeneity of HBV is known to be high. Based on phylogenetic analysis using a complete viral genome, HBV is classified into 10 genotypes (A to J) and 35 sub-genotypes. The classification of genotypes A to H is based on intergenotypic divergence of >8 per cent in the whole genome. While A, C and D are the predominant genotypes, I and J were recently reported4. Distribution of HBV genotype/s varies from region to region. Genotype A is predominantly found in Africa, India, Europe and America, whereas genotype B and C are mostly detected in the Asia-Pacific region. Genotype D is found in India, Africa, Mediterranean regions and Europe. Most of the genotypes reported from West Africa, South and Central America are E and F respectively. Genotype I was detected in Vietnam, Laos and eastern parts of India and Genotype J from Japan4,5.
Intergenotypic recombination is known to be a common phenomenon in HBV and plays an important role in its evolution6. Although the exact reason is unknown, recombination may occur due to co-infection of a host with multiple genotypes or sub-genotypes of HBV6. There are some reported instances of HBV recombinant forms, such as B2-B5 formed by recombination between B and C genotypes and found in South-East Asia7; A and D genotype recombination reported from South Africa and India8 and C and D genotype recombination reported mainly from China9. There are two forms of the C/D recombinant; CD1 has D genotype sequence up to 799 bases, whereas CD2 has a large portion of D genome, up to 1499 bases9. Recombination between A and C, A and E and A and G genotypes has also been reported from other parts of the world10,11.
In India, genotype D is more prevalent, followed by genotype A. However, other genotypes, such as B, C, E, F and G, are also reported2. A novel recombinant designated as D9 sub-genotype of HBV with discrete recombination with genotype C at the pre-core and core region was reported from Eastern India12; however, data on recombinant genotypes of HBV from India are scarce. HBV is also classified into subtypes. HBV surface antigen (HBsAg) subtypes, such as adw2 and ayw1, are identified from the eastern, southern and north-eastern regions of India13. HBV genotype C is always associated with subtype adr and is found in the eastern and north-eastern regions. Subtypes adw3 and ayw2 are associated with genotype D and are found in the eastern and western regions, whereas ayw3 spreads throughout, including south, east, north-east and western parts of India13.
Although India is designated as an intermediate endemic country for HBV, there are several pockets with a high prevalence of HBV, of which many are tribal-dominated areas14. Recently, we had conducted a community-based cross-sectional study in the scheduled districts of Lahaul and Spiti in Himachal Pradesh, India, and documented a high prevalence of HBV15. A genotyping study of HBV from the Spiti valley by Sharma et al16 identified genotypes C and D in circulation in the population. However, there is limited information about HBV serotypes, genotypes and sub-genotypes circulating in Lahaul and Spiti valleys. Moreover, infecting genotype is also considered one of the risk factors in HBV infection. Hence, to fill this lacuna, this genotyping study was conducted on HBsAg-positive samples collected from this area.
Material & Methods
The present study was conducted at the division of Virology and Zoonosis, Indian Council of Medical Research-National Institute of Research in Tribal Health, Jabalpur, Madhya Pradesh. This study was approved by the Institute Ethical Committee (Letter No. NIRTH/IEC/3895/2017).
Study area: The Lahaul and Spiti district is situated 13,000 feet above sea level in the Himalayan ranges in Himachal Pradesh, India. It has extreme weather conditions, with average snowfall up to seven feet and temperatures dropping to −30°C in winter, closing down almost all non-emergency services (https://hplahaulspiti.nic.in/). A seroprevalence study conducted previously to estimate HBV prevalence and its risk factors in 32 villages of the district showed a high HBV prevalence of 10.5 per cent (141/1339)15. The prevalence of HBV was higher in Spiti valley (17.3%) compared to Lahaul valley (3.9%). We conducted this molecular study on 14 HBsAg-positive samples available in sufficient quantity for molecular testing. The samples were tested for the presence of HBsAg by ELISA from March to October 201715, and the molecular studies were conducted from December 2019 to June 2020.
Amplification of surface gene and reverse transcriptase region: HBsAg ELISA-positive serum samples were subjected to DNA isolation using QIAamp DNA Mini Kit (Quigen, Hilden, Germany ) following the manufacturer’s protocol. Amplification of partial surface gene and reverse transcriptase region (972 bp) was carried out using primer sequence described by Arankalle et al, 201117; the PCR kit used had proofreading activity (Invitrogenby Life Technologies, Waltham, MA, USA). The PCR-amplified products were loaded onto 1.5 per cent agarose gel for electrophoresis, and products of 972 bp band size were gel excised. DNA was purified from the excised gel using HiYield Plus™ Gel Extraction Kit (Real Biotech Corp., Taiwan) following the manufacturer’s protocol. Purified PCR products were sequenced using BigDye Terminator Cycle Sequencing Kit on ABI 3130 XL genetic analyzer (both from Applied Biosystems, USA).
Sequence analysis: The nucleotide sequences obtained were quality checked, curated manually and analyzed for homology using the Basic Local Alignment Search Tool (BLAST).
Phylogenetic analysis: To plot the phylogenetic tree, 45 highly homologous HBV reference sequences were selected from GenBank for each genotype. Further reference sequences for genotype D [n=18 (sub-genotype D3, n=6; D1, D2 and D5, n=2 each and D4, D6, D7, D8, D9 and D10, n=1 each)], genotypes A and B (n=2 each), genotype C (n=4), E to I (n=2 each), genotype J (n=1), CD1 (n=5) and CD2 (n=3) were retrieved from GenBank database and aligned using software ClustalW. A phylogenetic tree was constructed by employing the neighbour-joining method in MEGA software version 5.05 by applying 1000 bootstraps to determine the circulating genotype/s of HBV18 (Fig. 1).
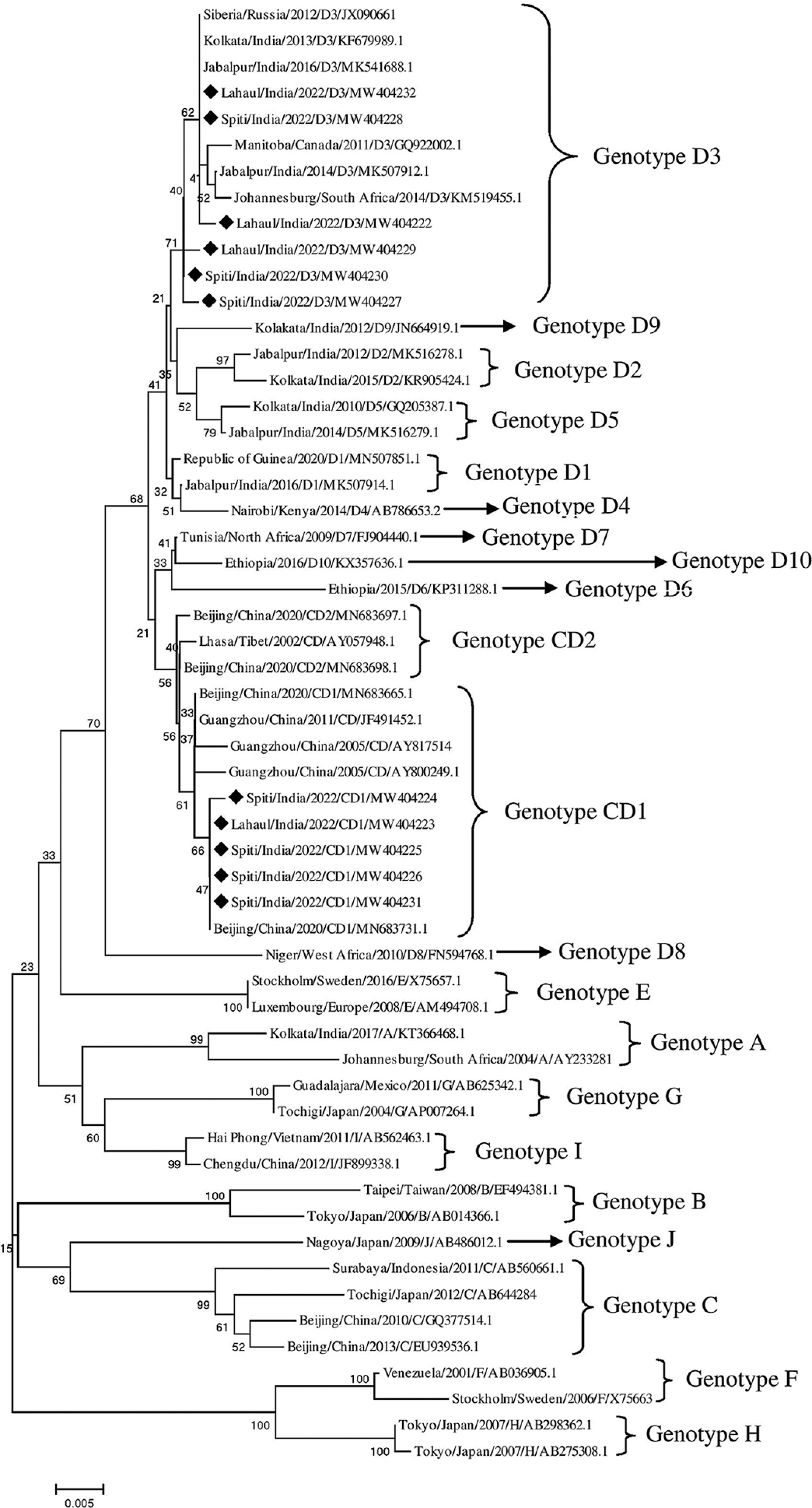
- The phylogenetic tree of HBV sequence detected from Lahaul and Spiti (marked with ♦) was constructed using the neighbour-joining method in MEGA5. The tree was constructed by using reference sequences for genotype D (n=18), genotypes A and B (n=2 each), genotype C (n=4), E to I (n=2 each), genotype J (n=1), CD1 (n=5) and CD2 (n=3) were retrieved from GenBank database. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Pattern of labelling: City/Country/Year/Genotype/Accession Number).
Recombination analysis: The sequences were subjected to recombination analysis using the jumping profile Hidden Markov Model (JPHMM) using an online tool (http://jphmm.gobics.de/), as described by Schultz et al19, and circular figures were plotted (Fig. 2).

- Genome recombination analysis in (A) HBV/CD isolate (MW404226); (B) HBV/D isolate (MW404222) by schematic diagram of jpHMM results showing the circular HBV genome with genotype CD and genotype D regions. The outer ring shows the predicted genotype recombination. The yellow colour represents genotype D whereas genotype C is marked in green colour of HBV.
Subtyping of hepatitis B virus: Nucleotide sequences of the S gene were translated in silico using the ExPASy - Translate tool. Subtyping of HBV was done according to the amino acid sequences at codons 122, 127, 134, 140, 159 and 160 of the S region20.
Results
To access the genetic variation of HBV in Lahaul and Spiti, we amplified the partial S gene and rt-region (972 bp) of previously collected HBsAg-positive samples and sequenced them. We successfully sequenced 11 out of the 14 selected HBsAg-positive samples, and after curation, the sequences were submitted to GenBank (Accession No. MW404222-MW404232). Of these 11 serum samples, eight were from Spiti and three were from Lahaul. BLAST analysis of these sequences revealed that the circulating HBV belonged to at least two genotypes, genotypes D and CD. Six sequences belonged to genotype D, namely MW404222, MW404232, MW404227, MW404228, MW404229 and MW404230, which showed sequence similarity of 99.25, 99.35, 99.72, 99.72, 99.27 and 99.74 per cent, respectively, with MK541688.1. Of these six, two were from Lahaul and four were from Spiti. The other five sequences belonging to genotype CD, namely MW404223, MW404224, MW404225, MW404226 and MW404231, showed sequence similarity of 99.86, 99.72, 99.66, 99.89 and 100 per cent, respectively, with MN683731.1 (CD1); four were from Spiti and one from Lahaul. Further sub-genotyping revealed that those belonging to genotype D were of sub-genotype D3 and the rest five sequences belonged to sub-genotype CD1 (Fig. 1). The sequences of CD1 sub-genotype showed close homology with the sequences from China.
To confirm the intergenotypic recombination, we analyzed the sequences using jpHMM, a graphical display sequence analysis tool and found that there were two genotypes, D and CD, of viruses circulating in the area. It further confirmed that CD1 was formed by the integration of 806 (±27) nucleotides of genotype D into genotype C (Fig. 2). It was proposed that the CD1 sub-genotype should be called as recombino-subgenotype and not a separate sub-genotype21. The serotype/subtyping of all the 11 samples was found to be ayw2.
Discussion
This study reports recombinant genotypes CD1 and D3 in the Himalayan Lahaul and Spiti valleys of India, which has a high prevalence of HBV and shares its border with Tibet. A, C and D are the most commonly found HBV genotypes in India; geographically, A and D are predominantly found in Northern and Southern India, whereas C is predominant in the eastern part of India2. The reason behind the diverse HBV genotype distributions in India is its geographical presence between West, Central and East Asian countries. This along with the genetic, and sociocultural diversity of the Indian population given the anthropological migrations is thought to contribute to the gene flow from neighbouring countries. This multi-ethnicity of the Indian population is also reflected in the distribution of HBV genotype across different parts of the country20. Additionally, the recent increase in trade and the frequent physical communication between the countries has also influenced circulation of HBV genotypes.
HBV infection causes innocuous to life-threatening fulminant disease. The asymptomatic HBV sometimes progresses to cirrhosis and hepatocellular carcinoma. There are studies showing that HBV genotype are directly linked with disease severity, pathogenesis and antiviral based treatment efficacy5.
In the Taiwanese population, genotype C was responsible for the higher preponderance of HB e-antigen (HBeAg)-positive chronic infection22, whereas genotypes B and C were responsible for infection in children23. A Spanish study showed an association between the reduction of HBeAg seroconversion in genotype A patients. Further, the genotype A- and B-infected patients have higher HBsAg seroclearance compared with C- and D-infected patients. The wild-type genotype E reportedly has a tendency for advanced infection in Gambian-vaccinated children24.
In our previous study15 a high prevalence of HBV infection was reported in Lahaul and Spiti district of Himachal Pradesh, wherein Spiti valley recorded 17.2 per cent HBsAg positivity. High-risk sexual behaviour, family history of HBV positivity and occupations such as student and preacher were the most prominent social risk factors15.
In the present study, molecular analysis using different tools revealed that HBV genotypes in Lahaul and Spiti are of two major categories. Of the 11 sequences, six belonged to genotype D and five belonged to CD1 recombinant genotype. The detection of genotype D in the area is in line with the reports published by other investigators in mainland India. Genotype D is predominant in the northern, western and southern parts of India. It is also distributed around the world, mainly in Saudi Arabia, Iran, Afghanistan, Pakistan and Turkey. Genotype D is further divided into different sub-genotypes and follows a pattern of geographical distribution4. In the present study, we detected sub-genotype D3 from the area. Sub-genotype D1 is prevalent in the Mediterranean region and D2, D3, D4, D5, D6, D7 and D8 are found in Africa, Eastern Europe, Australia, Indonesia and India. Interestingly, several published documents have described intergenotypic recombination of genotype D with newly obtained or existing genotypic variants, such as A or C25.
Intergenotypic recombination is an important phenomenon in HBV genetic variability and it has been reported globally. Further, it has been found that recombinant forms are spreading within the human population and developing their specific distributions globally. Published data have revealed that recombination between two different genotype strains develops mosaic HBV genomes. Worldwide, different recombinations have been identified, such as A/B, A/C, A/E, C/E, C/D, D/A, D/E and D/E/A26. Knowledge of the circulating genotype, recombination and mutation patterns of HBV can help clinicians predict the prognosis of the disease and decide treatment modalities.
We detected the CD1 recombinant sub-genotype in the Lahaul and Spiti region; however, Sharma et al16 using pre-S1/S2 and polymerase genes reported a high prevalence of genotype C, followed by genotype D. Similarly, Khan et al27 reported the dominance of HBV genotype D, followed by genotypes C and A in the adjoining Ladakh region. Lahaul and Spiti district is bordered by Tibet and previous studies have shown the dominance of HBV sub-genotype CD1 in the adjoining Tibet area and Western China, mainly in the Qinghai-Tibet plateau9,28. Furthermore, Borkakoty et al29 have found that genotype A, C, D and C/D hybrids are common in the tribal population of Arunachal Pradesh, who had migrated from Tibet. A retrospective study from Arunachal Pradesh, another State bordering Tibet, showed the presence of genotypes A, C, D and G; however, the full-genome sequencing of genotype G revealed that it was in fact a novel recombinant genotype I30.
Two types of HBV/CD recombinants have been reported earlier, HBV/CD1 (10-800 nucleotides from genotype D integrated into genotype C) and HBV/CD2 (10-1500 nucleotides from genotype D integrated into genotype C)28. CD recombinant genotype was derived from the sub-genotype C2 and genotype D and is prevalent in western China28.
Although recombination is well documented in HBV and more than 30 recombinants are reported, their nomenclature, taxonomic position and classification are debatable. Pourkarim et al21 suggested a terminology ‘recombino-subgenotype’ instead of calling them an independent sub-genotype. Conducting full-genome sequencing on a large number of samples and conducting in-depth bioinformatic analyses on the samples collected from the area can aid in further classification.
It is speculated that the possible geographic origin of the CD recombinant is the Silk Road, a transcontinental trade route that linked China in the East with Europe in the West31. This recombination may be due to the geography and genetic background of the population. It is possible that this CD1 recombinant might have migrated with the Tibetan population to Lahaul and Spiti as they share common geographical, cultural and religious beliefs28.
The present study had a few limitations, such as limited number of samples, absence of full-genome sequencing and next-generation sequencing (NGS) data and absence of patient follow up clinical outcome data. Nonetheless, this study reports the presence of the CD1 recombinant along with genotype D3 in Lahaul and Spiti in Himachal Pradesh, India, which may be due to past or present transcontinental migration between the adjacent countries. Further, a detailed study to understand the prevalence of the CD recombinant, its clinical implications and full-genome sequence using NGS and mutational analysis will help understand the origins of this recombinant and to combat the disease.
Acknowledgment:
The authors acknowledge Dr Naveen Minahs, Mr. M.K Shukla and staff of Keylong Field Station of ICMR-NIRTH for their active participation in the study. We also thank Dr Sri Krishna for help in sequence analysis.
Financial support & sponsorship: The study received financial support from Indian Council of Medical Research, New Delhi. (DHR-ICMR: Grant No. VIR/43/2011-ECD-I and No. DHR/VDL/04/2018) and ICMR-NIRTH, Jabalpur (intramural grant).
Conflicts of Interest: None.
References
- World Health Organization. Hepatitis. Available from: https://wwwwhoint/news-room/fact-sheets/detail/hepatitis-b
- Molecular characterization of hepatitis B virus reveals circulation of multiple subgenotypes of genotype D with clinically important mutations in central India. Indian J Med Microbiol. 2021;39:67-72.
- [Google Scholar]
- Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-29.
- [Google Scholar]
- Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141-50.
- [Google Scholar]
- Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436.
- [Google Scholar]
- Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467-76.
- [Google Scholar]
- Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology. 2003;124:925-32.
- [Google Scholar]
- Identification and characterization of genotype A and D recombinant hepatitis B virus from Indian chronic HBV isolates. World J Gastroenterol. 2008;14:6228-36.
- [Google Scholar]
- Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet plateau. PLoS One. 2011;6:e18708.
- [Google Scholar]
- A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047-56.
- [Google Scholar]
- Hepatitis B virus genotype G epidemiology and co-infection with genotype a in Canada. J Gen Virol. 2008;89:3009-15.
- [Google Scholar]
- New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013;20:209-18.
- [Google Scholar]
- Molecular epidemiology and genetic characterization of hepatitis B virus in the Indian subcontinent. Int J Infect Dis. 2014;20:1-10.
- [Google Scholar]
- Sytematic review and meta-analysis of prevalence of hepatitis B in India. Indian Pediatr. 2007;44:663-74.
- [Google Scholar]
- Seroprevalence and risk factors of hepatitis B virus infection in tribal population of Himalayan district Lahaul and Spiti, India. Pathog Glob Health. 2019;113:263-7.
- [Google Scholar]
- Genotyping of hepatitis B virus isolates from Lahaul and Spiti district in Himachal Pradesh, India. Indian J Gastroenterol. 2018;37:261-5.
- [Google Scholar]
- An outbreak of hepatitis B with high mortality in India: Association with precore, basal core promoter mutants and improperly sterilized syringes. J Viral Hepat. 2011;18:e20-8.
- [Google Scholar]
- MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9.
- [Google Scholar]
- jpHMM: Recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res. 2012;40:W193-8.
- [Google Scholar]
- Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24-34.
- [Google Scholar]
- Molecular identification of hepatitis B virus genotypes/subgenotypes: Revised classification hurdles and updated resolutions. World J Gastroenterol. 2014;20:7152-68.
- [Google Scholar]
- Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J Clin Microbiol. 2002;40:1207-9.
- [Google Scholar]
- Breakthrough HBV infection in vaccinated children in Taiwan: Surveillance for HBV mutants. Antivir Ther. 2010;15:463-9.
- [Google Scholar]
- Envelope protein variability among HBV-Infected asymptomatic carriers and immunized children with breakthrough infections. J Med Virol. 2008;80:1537-46.
- [Google Scholar]
- A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J Gen Virol. 2005;86:985-90.
- [Google Scholar]
- Identification of hepatitis B virus genotype A/E recombinants in Ghana. Virus Genes. 2019;55:707-12.
- [Google Scholar]
- Epidemiology of hepatitis B and C viral infections in Ladakh Region. Indian J Gastroenterol. 2018;37:504-10.
- [Google Scholar]
- Whole-gene analysis of two groups of hepatitis B virus C/D inter-genotype recombinant strains isolated in Tibet, China. PLoS One. 2017;12:e0179846.
- [Google Scholar]
- Circulating genotypes of hepatitis B virus in Arunachal Pradesh. Indian J Med Res. 2008;127:65-70.
- [Google Scholar]
- Circulation of genotype-I hepatitis B virus in the primitive tribes of Arunachal Pradesh in early sixties and molecular evolution of genotype-I. Infect Genet Evol. 2014;27:366-74.
- [Google Scholar]
- Genotype D of hepatitis B virus and its subgenotypes: An update. Hepatol Res. 2013;43:355-64.
- [Google Scholar]






