Translate this page into:
Development of a RT-LAMP assay for detection of SARS-CoV-2
For correspondence: Dr Shyam Sundar Nandi, ICMR-National Institute of Virology, (Mumbai Unit), Haffkine Institute Compound, A. D. Marg, Parel, Mumbai 400 012, Maharashtra, India e-mail: nandibiotech@gmail.com
-
Received: ,
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
The pandemic of SARS-COV-2 began in Wuhan, China in December 2019 and has caused more than 101 million cases worldwide. Diagnostic technologies possessing sensitivity and specificity equivalent to real-time reverse-transcriptase polymerase chain reaction (rRT-PCR) assays are needed to ramp up testing capacity in most countries. Newer platforms need to be technically less demanding, require minimum equipment and reduce turn-around time for reporting results. The objective of this study was to exploit loop-mediated isothermal amplification (LAMP) for the detection of SARS-CoV-2 and evaluate its performance by comparison with rRT-PCR.
Methods:
Reverse-transcription LAMP (RT-LAMP) assay primers were designed to detect envelop (E) and nucleocapsid (N) genes of SARS-CoV-2. Positive control RNA was prepared by in vitro transcription of E and N genes clones. RT-LAMP amplification reactions were incubated at 65°C for 30 min. Results were recorded visually. RT-LAMP results were evaluated by comparing the results obtained with a commercial rRT-PCR kit.
Results:
The RT-LAMP assay for E and N genes was carried out in separate tubes. RT-LAMP detected about 40 copies of SARS-CoV-2 RNA per reaction. A total of 253 throat swabs were tested using the RT-LAMP assay. The overall diagnostic sensitivity and specificity of the LAMP assay were 98.46 and 100 per cent, respectively, as compared to the rRT-PCR.
Interpretation & conclusions:
SARS-CoV-2 RT-LAMP assay was designed, standardized and evaluated. The assay showed diagnostic sensitivity and specificity equivalent to rRT-PCR assays. The assay will be useful to increase testing capacity for the detection of SARS-CoV-2 in the country.
Keywords
COVID-19
isothermal amplification
RT-LAMP
SARS-CoV-2
visual detection
The pandemic of SARS-CoV-2 has emphasized a need for rapid screening of the population to track the spread. The current diagnostic strategy takes into account the clinical symptoms along with virus detection. The virus detection methods involved are molecular-based and immunoassays for SARS-CoV-2. Real-time reverse-transcription polymerase chain reaction (rRT-PCR) is a widely used test in India and worldwide. The rRT-PCR molecular assay is considered as a gold standard method for the detection of SARS-CoV-212. In addition, cartridge-based nucleic acid amplification test (CBNAAT)3 and TrueNat™ are the two molecular methods used as ‘point of care’ diagnostic for COVID-194. FNCAS9 editor linked uniform detection assay (FELUDA) is a clustered regularly interspaced short palindromic repeats (CRISPR) based assay which also is used for the detection of SARS-CoV-25.
Attempts have been made by various researchers to develop reverse transcription loop-mediated isothermal amplification (RT-LAMP)-based COVID-19 diagnostic assays using various detection methods such as fluorescent dye (SYBR green)6, labelled probes or turbidity due to precipitation of magnesium pyrophosphate7 and visual detection by colour change methods. Thi et al8 reported an RT-LAMP assay-based sequencing for the detection of SARS-CoV-2. This study was undertaken to develop a colorimetric endpoint RT-LAMP assay for diagnostic sensitivity and specificity equivalent to rRT-PCR assays for COVID-19.
Materials & Methods
This study was carried out from April to July 2020 at the ICMR-National Institute of Virology, Mumbai Unit (NIVMU), Mumbai, India. The NIVMU is an approved laboratory for testing COVID-19 samples. Clinical samples were received anonymously. The Institutional Ethics Committee has approved the manuscript. Nasal/throat swabs of suspected COVID-19 patients collected in the virus transport medium by various hospitals were received for laboratory diagnosis under COVID-19 surveillance. Samples were tested by real-time RT-PCR as per the ICMR standard protocol29 and results reported to the sender hospital, State government and the ICMR database. Samples were tested retrospectively by the RT-LAMP assay. The patient’s demographic information and clinical details were not provided to the laboratory for retrospective testing. Bio-safety laboratory practices were strictly followed for sample processing and testing.
Primer designing for LAMP assay: Genomic sequences of SARS-CoV-2, SARS-CoV and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) strains were retrieved from the NCBI website (https://www.ncbi.nlm.nih.gov/). The sequences were grouped according to the six WHO Regions. Multiple alignments were studied using MEGA-X software with the CLUSTAL-W algorithm (https://www.megasoftware.net/). SARS-CoV-2 envelope gene (E) and nucleoprotein gene (N) were chosen for RT-LAMP assay development.
RT-LAMP primers were designed using the online software PrimerExplorerV5 (https://primerexplorer.jp/e/ Eiken Chemical Co., Japan). The RT-LAMP primers set contained (i) F3/ B3 (Forward/ Backward outermost primers) which are similar to PCR primers, (ii) FIP/BIP (Forward/Backward inner primers), FIP and BIP are specialized primers with complementary and non-complementary nucleotide segments and (iii) LF/BF (Forward/Backward loop primers). The primers were custom synthesized by SIGMA Laboratories, USA.
Positive test control: The E and N genes of SARS-CoV-2 were cloned in TA cloning vector pTZ57R/T (Thermo Fisher Scientific, USA. Cat No. K1213). The cloning primers were designed with 5’ T7 Promotor sequence overhang. In vitro transcription of the cloned fragment was done using T7 RNA polymerase (Thermo Fisher Scientific, USA Cat No. K0441). RNA transcripts were purified using GeneJet RNA Clean-up and Concentration Micro kit (Thermo Fisher Scientific, USA Cat No. K0842). The concentration of RNA in positive template control (PTC) of E and N genes was 760 ng/µl each. Ten-fold serial dilutions of the RNA transcripts were used for calculating concentration and copy numbers by Qubit™ RNA HS Assay kit (Thermo Fisher Scientific, USA Cat. No. Q32852).
RT- LAMP assay:
Primer mix stock: The N gene LAMP primer set contained F3/B3, FIP/BIP and both forward and backward loop primers. The E gene set contained F3/ B3, FIP/BIP and only the backward loop primer. Stocks (10X) of primer mixtures were prepared separately for the E and N genes in nuclease-free water and stored at −20°C. The 10X stock contained 2 µM of F3 and B3; 16 µM of FIP and BIP, and 4 µM of LF/LB in nuclease-free water.
RT-LAMP reaction set-up: For every test run, the RT-LAMP reaction master mix was prepared freshly for the number of samples being tested. The master mix contained 2µl of 10X primer mix (E and N genes in separate tubes), 10 µl WarmStart Colorimetric LAMP 2X Master Mix with uracil DNA glycosylase (UDG) (New England BioLab, Ipswich, MA, USA Cat No. M1804L), 0.8 µl Enhancer solution and 3.2 µl of nuclease-free water per sample tested. A volume of 16 µl of the master mix was distributed in 200 µl PCR tubes. The reaction mixture was vortexed and spun down. It was ensured that the reaction solution had a bright pink color, which indicated the initial high pH required for effective RT-LAMP reaction. Non-template controls (NTC) and positive controls (E and N genes) were included in each run; 4 µl of nuclease-free water was added in NTC, 4 µl of template RNA and 4 µl of positive control RNA in respective RT-LAMP reaction tubes.
The reaction tubes were incubated at 65°C for 30 min in a Thermal Cycler (Thermo Fisher Scientific, USA). The results were recorded by visual reading of the reaction colour. A change of colour from pink to yellow indicated a positive reaction.
RNA extraction: RNA was extracted from 140 µl of the samples using QIAamp Viral RNA Mini Kit (QIAGEN Germany, Cat No: 52906) as per the manufacturer’s instructions. RNA was eluted in 30 ul of Elution buffer (AVE; Qiagen) provided with the kit.
Reference standard assay: A real-time RT-PCR assay was used as the reference method for comparative evaluation of the RT-LAMP assay. LabGun COVID-19 PCR Kit (Lab Genomics Co., Ltd., R, Cat No. CV9017, Republic of Korea), approved for COVID-19 diagnosis by the Ministry of Health was used as per the manufacturer’s instructions. This assay tests for RdRp gene (SARS-CoV-2) and E gene (Sarbecovirus). The assay recommended cycle threshold value (Ct) ≤40 for sample positivity. Amplification of test RNA by RdRp-specific primers was confirmatory of COVID-19 infection.
Interpretation of RT-LAMP and rRT-PCR assays: The RT-LAMP endpoint was the visual recording of the colour of the reaction. The change of colour of the reaction from pink to yellow indicated a positive reaction. For a test run to be valid, the PTC tube should become yellow, the NTC should remain pink. Test sample results were recorded for the E and N genes separately.
rRT-PCR results were interpreted as per the manufacturer’s instructions. Ct values ≤40 were interpreted as a positive test. Samples showing RdRp gene negative and E gene-positive reactions were interpreted as inconclusive. RdRp gene amplification was specific (confirmatory) for SARS-CoV-2 even in the absence of E gene positivity.
Statistical analysis: The screening test evaluation module of OpenEPI software (https://www.openepi.com/) was used for statistical analysis. The statistical analysis was performed considering the rRT-PCR as a gold standard assay. The sensitivity and specificity of RT-LAMP assay were calculated by comparing positivity of individual gene (either E or N gene) as well as both genes positivity criteria. The results with 95 per cent confidence interval are included.
Results
RT-LAMP: Initially, multiple sets of RT-LAMP primers were generated. Four key criteria, namely (i) melting temperature (Tm), (ii) stability at the 3’ and 5’ ends (Delta G), (iii) GC content, and (iv) possibility to form secondary structures were used for further narrow down. Primers were designed so that their GC contents were about 50-60 per cent. The Tm values were between 64 and 66°C for F1c and B1c, 59 and 61°C for F2, B2, F3, and B3, and 64 and 66°C for the loop primers. The 3’ ends of F2/B2, F3/B3 and the 5’ end of F1c/B1c were designed, had free energy −4 kcal/mol or less. The selected sets of the RT-LAMP primers for E and N genes are shown in Table I and their relative locations on the SARS-CoV-2 genome in Figure 1A and B. E gene assay had five primers including one loop primer located between nucleotides 26231 and 26450. The set of the N gene assay had six primers including two loop primers located between nucleotides 28280 and 28499. The sequence alignments (Fig. 1A and B) showed that the E gene primers F3, F1c and LB had zero mismatches; F2, B1c and B3 primers each had one mismatch and the B2 primer three mismatches with sequences of the other β coronavirus consensus sequence. Similarly, alignment of N gene primers revealed that the F3 primer had three gaps and four mismatches; B1c six mismatches; LF, F2, F1c and B3 two mismatches and LB primer had no mismatch.
| Sequence ID | 5’ Sequence 3’ | Position | 5’ΔG | 3’ΔG | GC content | Tm (⁰C) |
|---|---|---|---|---|---|---|
| E gene primers | ||||||
| SUSTJ_ E-F3 | TGAGTACGAACTTATGTACTCAT | 26232-26254 | −3.99 | −4.29 | 56.33 | 54.7 |
| SUSTJ_ E-B3 | TTCAGATTTTTAACACGAGAGT | 26441-26420 | −4.27 | −4.59 | 55.31 | 57.1 |
| SUSTJ_ E-FIP | F1c: ACCACGAAAGCAAGAAAAAGAAGT- | 26295-26318 | −4.85 | −4.74 | 55.36 | 80.4 |
| F2: TCGTTTCGGAAGAGACAG | 26255-26272 | −6.33 | −4.24 | 61.55 | ||
| SUSTJ_ E-BIP | B1c: TTGCTAGTTACACTAGCCATCCTT- | 26323-26346 | −4.5 | −5.74 | 56.2 | 75.8 |
| B2: AGGTTTTACAAGACTCACGT | 26406-26387 | −4.49 | −4.24 | 61.42 | ||
| SUSTJ_E-LB | GCGCTTCGATTGTGTGCGT | 26350−26368 | −6.92 | −6.73 | 63.56 | 69.8 |
| N gene primers | ||||||
| SUSTJ_ N-F3 | CCCCAAAATCAGCGAAATGC | 28289-28308 | −5.96 | −4.56 | 59.82 | 68.4 |
| SUSTJ_ N-B3 | ATTGGAACGCCTTGTCCTC | 28494-28476 | −4.41 | −5.2 | 59.41 | 64.0 |
| SUSTJ_ N-FIP | F1c: GCCCCACTGCGTTCTCCATTC- | 28377-28360 | −3.39 | −5.69 | 59.52 | 87.1 |
| F2: CATTACGTTTGGTGGACCCT | 28315-28334 | −7.2 | −4.51 | 65.55 | ||
| SUSTJ_ N-BIP | B1c: GCGATCAAAACAACGTCGACCC- | 28378-28399 | −5.85 | −4.76 | 59.51 | 87.6 |
| B2: CCTTGCCATGTTGAGTGAGA | 28457-28438 | −5.93 | −6.63 | 64.17 | ||
| SUSTJ_ N-LF | TGGTTACTGCCAGTTGAATCTG | 28356-28335 | −4.25 | −3.90 | 60.29 | 63.9 |
| SUSTJ_ N-LB | GGTTTACCCAATAATACTGCGTCTT | 28403-28427 | −3.85 | −5.18 | 61.12 | 63.8 |
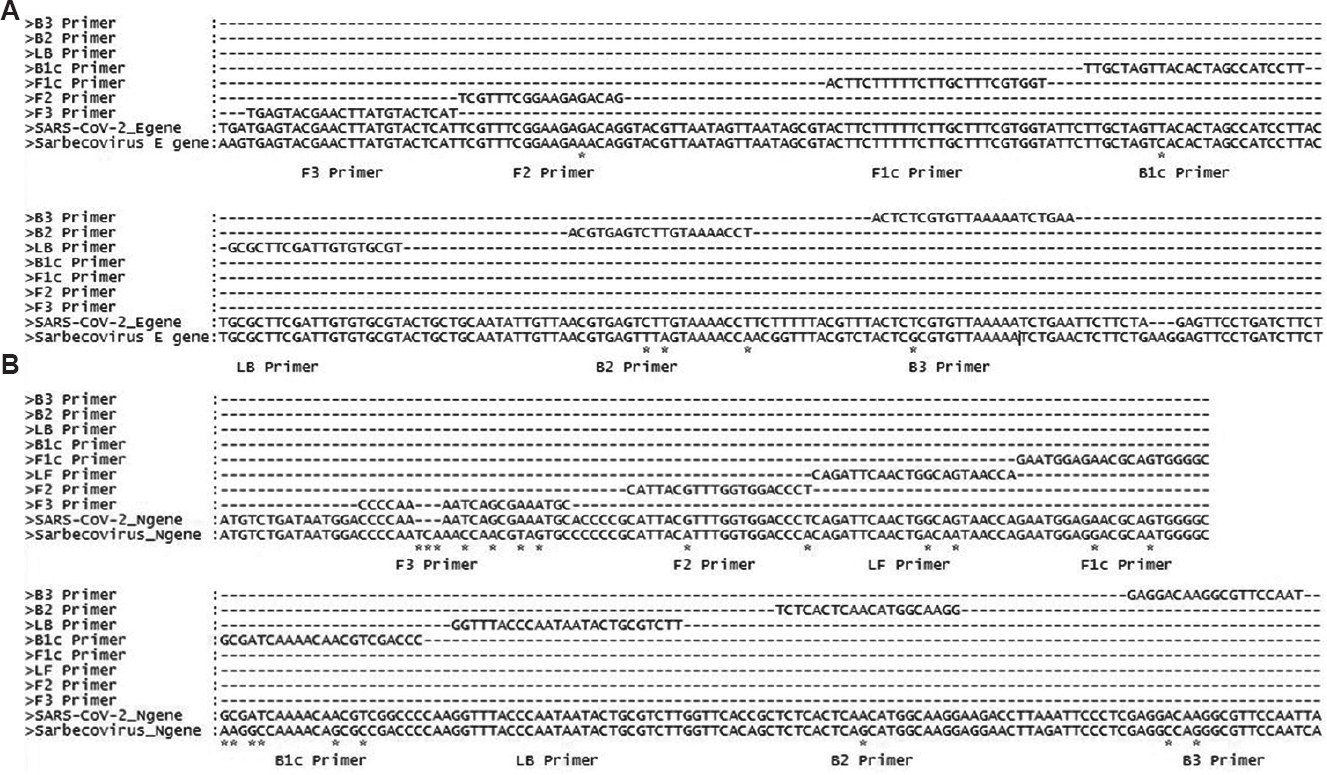
- (A) Alignment of E gene of SARS-CoV (NC_004718.3) and SARS-CoV-2 (NC_045512.2) reference sequences showing primer positions and highlighting mismatches using ‘*’. F3, F1c and LB primers with no mismatches; while F2, B1c and B3 primers having one mismatch and B2 primer showing three mismatches. (B) Alignment of N gene of SARS-CoV (NC_004718.3) and SARS-CoV-2 (NC_045512.2) reference sequences showing primer positions and highlighting mismatches using ‘*’. F3: having three gaps and four mismatches; B2: showing one mismatch; LF, F2, F1c and B3: showing two mismatches; B1c: showing six mismatches and LB has no mismatch.
A single temperature (isothermal) for incubation makes RT-LAMP assay easy to set up and visual recording of the results without ambiguity. The E and N genes RT-LAMP reactions were carried out in separate tubes. Fig. 2 shows colour change from pink to yellow after amplification of the SARS-CoV-2 RNA template. The possible outcome of RT-LAMP reaction for a sample could be both genes negative, only E gene positive, only N gene positive and, both E and N genes positive. In case a sample exhibits only E gene positive or only N gene positive or both E and N genes positive in all the above scenarios, the sample is considered as positive for SARS-CoV-2 RNA. However, in the case of a sample, if both the genes display negative results, the sample is considered as negative for SARS-CoV-2 RNA (Fig. 2). By the virtue of the specifically designed primers, it was established that the sample showing single gene positivity to be considered as a positive result for SARS-CoV-2 RNA.
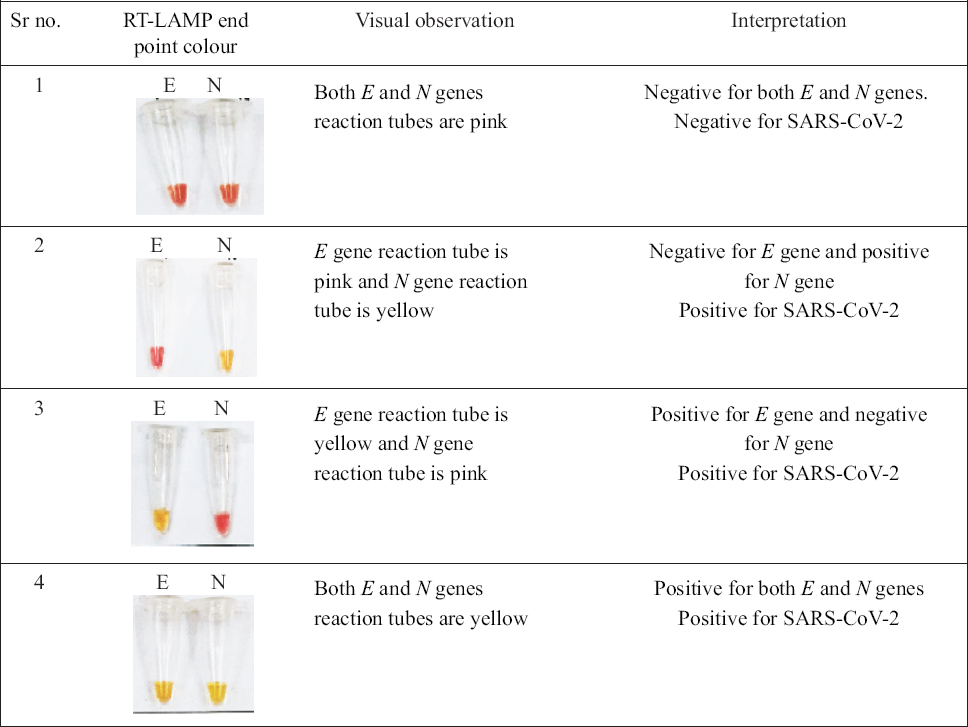
- Interpretation of results for detection of SARS-CoV-2 RNA: If the sample exhibits only E gene positive or only N gene positive or both E and N genes positive in all the above scenarios, the sample is considered as positive for SARS-CoV-2 RNA. However, in case of a sample, if both the genes display negative results, the sample is considered as negative for SARS-CoV-2 RNA.
Limit of detection: The limit of detection was calculated by performing the ten-fold serial dilutions of positive controls for each gene. The positive control RNA was obtained by performing in vitro transcription of cloned fragments of E and N genes in TA vector. The concentration of each dilution was calculated by the Q-bit method and according to the concentration the copy number was calculated 2.83×1011 for E gene and 3×1011 for N gene. The limit of detection was calculated to be 10 copies/µl. This corresponded to 40 copies per RT-LAMP reaction as the volume required per reaction was 4 µl (Supplementary Table I).
| Criteria | RT-LAMP | rRT-PCR |
|---|---|---|
| Simplicity | Easy to set up the reaction as it involves mixing of only three components. The reaction can be set up at room temperature also, as the enzymes used in this reaction are protected by Aptamer and can only be activated at temperature above 45°C. | Requires complicated reaction set up. The fluorescent dyes are light sensitive so the reaction has to be set up in a dark environment without exposure to direct light. Also the reaction requires cool packs during set up. |
| Time | RT-LAMP requires 30 to 40 min only | Usually requires to 1 to 2.5 h to complete the reaction |
| Temperature conditions | RT-LAMP requires isothermal temperature of 65°C throughout the reaction. | RT-PCR requires three conditions which are denaturation, annealing and extension working at different temperatures and times. This is possible only by using a thermal cycler. |
| Result interpretation | The results are interpreted by naked eye by observing the color change. No sophisticated instrument and trained technical staff is required to interpret the results. | The results are interpreted by measuring fluorescent emission. This requires sophisticated real-time PCR machines and also well trained technical staff. |
| Cost | RT-LAMP is comparatively cheaper. | rRT-PCR is expensive. |
| Instrument required | A simple heating block with accurate 65±0.5°C temperature can be used. | Sophisticated real-time PCR machines are required. |
Performance evaluation of RT-LAMP: RNA extracted from 253 clinical samples (throat swabs) were tested by the reference standard rRT-PCR targeting E and RdRp genes. RdRp gene results were regarded as confirmatory for the presence of SARS-CoV-2 RNA. Fig. 3 shows the distribution of Ct values of the RdRp gene of 134 samples, 119 samples showed Ct values >40 or no amplification (not included in Fig. 3). The 41 samples representing bar of high Ct values contained 15, 11, 7 and 8 samples in the Ct range 37, 38, 39 and 40, respectively.
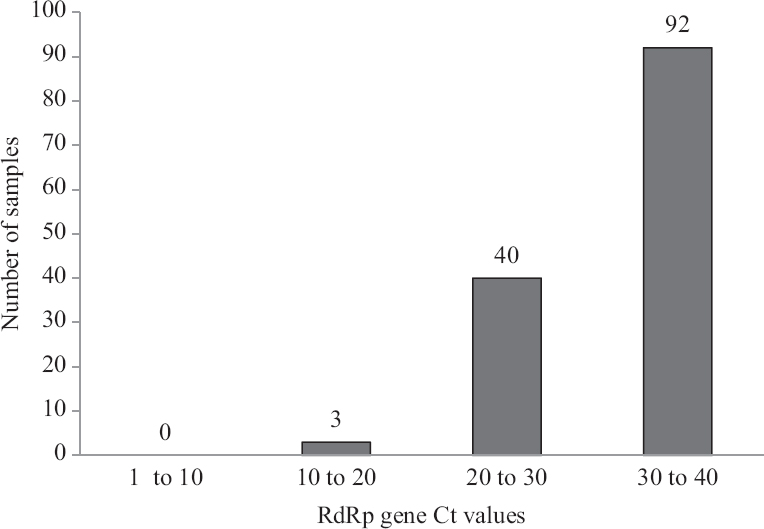
- The bar chart shows SARS-CoV-2 RdRp gene rRT-PCR cycle threshold (Ct) values of samples used for performance evaluation of the RT-LAMP assay. A total of 253 samples were tested, 119 samples with Ct >40 not included in the bar diagram.
The 253 samples were tested in RT-LAMP targeting E and N genes of SARS-CoV-2. Fourteen samples gave discordant results requiring detailed inquiry (Table II). As per the rRT-PCR kit manufacturer’s instructions the samples showing E gene positive and RdRp gene negative results were reported as inconclusive. All the 14 discordant samples showed E and RdRp genes Ct values very close to or above the maximum accepted for positivity (Ct=40). Four of the 253 samples were inconclusive. The four samples had Ct values >39 indicating borderline positivity for E gene. These samples were negative in both E and N genes RT-LAMP. Four samples were negative in E gene rRT-PCR with RdRp gene Ct values >39. These samples were negative in E and N gene RT-LAMP. Of the four samples negative in rRT-PCR, one was positive in both E and N gene RT-LAMP and three were positive for one of the two genes in RT-LAMP. In addition, two samples which were scored positive on the basis of RdRp/E gene Ct values (38.23, negative; and 38.77, 39.56), respectively, were scored E gene positive/N gene negative and, E and N genes negative in RT-LAMP. With due consideration, it was decided to modify the rRT-PCR result of the 4 ‘inconclusive’ samples to negative and four only RdRp (Ct>39) positive samples to negative for further analysis. The reference standard rRT-PCR assay thus detected 123 negative and 130 positive SARS-CoV-2 RNA samples.
| Sample ID | Ct value RdRp gene | Ct value E gene | rRT-PCR result | Modified interpretation of rRT-PCR results | RT-LAMP E gene | RT-LAMP N gene |
|---|---|---|---|---|---|---|
| 1 | 40.25 | 39.8 | Inconclusive* | Negative | Negative | Negative |
| 2 | 41.6 | 39.39 | Inconclusive* | Negative | Negative | Negative |
| 3 | UD | 39.82 | Inconclusive* | Negative | Negative | Negative |
| 4 | UD | 39.93 | Inconclusive* | Negative | Negative | Negative |
| 5 | 40.94 | UD | Negative | Negative | Positive | Positive |
| 6 | 40.09 | UD | Negative | Negative | Negative | Positive |
| 7 | 40.74 | UD | Negative | Negative | Positive | Negative |
| 8 | 40.56 | 40.61 | Negative | Negative | Positive | Negative |
| 9 | 38.23 | UD | Positive | Positive | Positive | Negative |
| 10 | 38.77 | 39.56 | Positive | Positive | Positive | Negative |
| 11 | 39.23 | 40.31 | Positive* | Negative | Negative | Negative |
| 12 | 39.74 | 40.81 | Positive* | Negative | Negative | Negative |
| 13 | 39.64 | UD | Positive* | Negative | Negative | Negative |
| 14 | 39.01 | 40.16 | Positive* | Negative | Negative | Negative |
* Samples with modified rRT-PCR interpretation. rRT-PCR, real time reverse transcription PCR; UD, undetectable; RT-LAMP, reverse transcription-loop mediated isothermal amplification
Table III presents comparative evaluation of results of RT-LAMP and real-time RT-PCR. E gene RT-LAMP reported 130 positives, 120 negatives and three PCR negatives as positives (false positive). Thus, E gene RT-LAMP showed 100per cent sensitivity and 97.56per cent specificity with the reference standard. N gene RT-LAMP reported 128 positives and 121 negatives (98.46% sensitivity and 98.37% specificity). Considering that a positive RT-LAMP reaction in either of the two genes (E or N) reports a SARS-CoV-2 positivity we got 100 per cent sensitivity with a slight loss of specificity (96.75%). Using a more stringent criterion that a sample should be positive in both the genes (E and N) for the confirmed presence of SARS-CoV-2 RNA, we got 98.46 per cent sensitivity and 100per cent specificity. The sensitivity and specificity of the RT-LAMP assay were thus found to be equivalent to real-time RT-PCR.
| Gene | Positive | Negative | Total tested | Sensitivity, % (95% CI) | Specificity, % (95% CI) | Positive predictive value (95% CI) |
|---|---|---|---|---|---|---|
| E gene | ||||||
| Positive | 130 | 3 | 133 | 100% (97.13-100) | 97.56% (93.58-99.07) | 97.74% (93.58-99.23) |
| Negative | 0 | 120 | 120 | |||
| Total | 130 | 123 | 253 | |||
| N gene | ||||||
| Positive | 128 | 2 | 130 | 98.46% (94.56-99.58) | 98.37% (94.26-99.55) | 98.37% (94.56-99.58) |
| Negative | 2 | 121 | 123 | |||
| Total | 130 | 123 | 253 | |||
| Either E or N gene | ||||||
| Positive | 130 | 4 | 134 | 100% (97.13-100) | 96.75% (91.94-98.73) | 97.01% (92.58-98.83) |
| Negative | 0 | 119 | 119 | |||
| Total | 130 | 123 | 253 | |||
| E and N genes | ||||||
| Positive | 128 | 0 | 128 | 98.46% (94.56-99.58) | 100% (96.97-100) | 100% (97.0-100) |
| Negative | 2 | 123 | 125 | |||
| Total | 130 | 123 | 253 |
CI, confidence interval
| Dilution | Concentration ng/µl | Copy number | Results |
|---|---|---|---|
| 1 | Too high | - | -ve |
| 10-1 | 76 ng/µl | 2.83×1011 | +ve |
| 10-2 | 12.5 ng/µl | 4.64×1010 | +ve |
| 10-3 | <20 ng/µl | 3.72×109 | +ve |
| 10-4 | <20 ng/µl | 108 | +ve |
| 10-5 | <20 ng/µl | 107 | +ve |
| 10-6 | - | 106 | +ve |
| 10-7 | - | 105 | +ve |
| 10-8 | - | 104 | +ve |
| 10-9 | - | 103 | +ve |
| 10-10 | - | 102 | +ve |
| 10-11 | - | 101 | +ve |
| 10-12 | - | 1 | -ve |
| 10-13 | - | 0.1/0 | -ve |
| 10-14 | - | 0.01/0 | -ve |
| 10-15 | - | 0.001/0 | -ve |
| Dilution | Concentration ng/µl | Copy number | Results |
|---|---|---|---|
| 1 | Too high | - | -ve |
| 10-1 | 76 ng/µl | 3×1011 | +ve |
| 10-2 | 12.7 ng/µl | 5.01×1010 | +ve |
| 10-3 | <20 ng/µl | 7.88×109 | +ve |
| 10-4 | <20 ng/µl | 108 | +ve |
| 10-5 | <20 ng/µl | 107 | +ve |
| 10-6 | - | 106 | +ve |
| 10-7 | - | 105 | +ve |
| 10-8 | - | 104 | +ve |
| 10-9 | - | 103 | +ve |
| 10-10 | - | 102 | +ve |
| 10-11 | - | 101 | +ve |
| 10-12 | - | 1 | -ve |
| 10-13 | - | 0.1/0 | -ve |
| 10-14 | - | 0.01/0 | -ve |
| 10-15 | - | 0.001/0 | -ve |
Discussion
The SARS-CoV-2 (Accession number NC_045512.2) specific conserved regions were identified for primer designing. Unlike the real-time PCR assays, RT-LAMP uses multiple forward and reverse (backward) primers and one or two loop primers. LAMP uses the RTx reverse transcriptase that gets activated >45°C and modified Bst 2.0 DNA polymerase having strand displacement activity1011. RT-LAMP reactions can therefore be carried out at a single incubation temperature (isothermal). Primers B2 and B3 initiate cDNA synthesis. FIP and BIP are specialized primers consisting of two parts, F2/F1c and B2/B1c, respectively. F2 and B2 bind with the template strand to initiate the amplification process while F1c and B1c sequences serve as overhangs which help loop formation as RT-LAMP reaction continues. The short distance between the F2 and F1c (and B2/B1c) helps the formation of a loop structure within the amplicon. The loop primers increase the number of initiation points for DNA synthesis by binding complementarily to the single-stranded loops and increase the pace of amplification12.
Unlike RT-PCR which requires denaturation, annealing and extension at different temperatures, the RT-LAMP reactions are incubated at a single temperature (isothermal reaction) usually at 65°C for 30 min. In the previously developed RT-LAMP assay for the detection of SARS-CoV-2 fluorescent-based detection system was used. This detection system has the advantage of multiplexing using Cas13 protein, but it requires ultraviolet trans-illuminator for result interpretation13. In the current study, the colorimetric RT-LAMP was developed which uses pH-sensitive dye phenol red. The results are read by the naked eye as the change of the color of the reaction mixture at the end of the incubation (pink to yellow) without the aid of any equipment. Another advantage is because it is an isothermal reaction so any incubation device which can maintain constant temperature (65°C) can be used. Therefore, LAMP assays are easy to perform, rapid and may become cost-effective.
Comparative testing of the newly developed RT-LAMP with real-time RT-PCR confirmed high sensitivity and specificity of the assay. The E and N gene primers possess multiple mismatches with other coronaviruses RNA sequences. For the LAMP assays, the FIP and BIP primers are responsible for complete specificity for amplification1415. Samples which were E gene positive/RdRp negative in rRT-PCR were all negative in our RT-LAMP assay confirming E gene specificity of the RT-LAMP assay. As per the instructions given by the manufacturer of the rRT-PCR used as reference assay, E gene positivity should be considered as an indication of a Sarbecovirus and the RdRp gene amplification was necessary for confirmation of SARS-CoV-2.
There were four samples which were negative for E gene amplification in rRT-PCR and positive for RdRp gene (Ct 39 to 40). The four samples were negative for both E and N genes in RT-LAMP. Whether clinical samples that were negative for Sarbecovirus E gene PCR and borderline positive for RdRp gene should be regarded positive for COVID-19 can be decided only by comparing clinical disease in the patients. These four samples were classified as negative for SARS-CoV-2 RNA for the analysis in the present study. The N gene primers contained various mismatches and at least one primer (F3) contained three nucleotides deletion regions providing high specificity of amplification in our RT-LAMP assay. Results confirmed that individually E gene RT-LAMP had 100 per cent sensitivity and N gene RT-LAMP sensitivity and specificity exceeded 98 per cent. Using more stringent criteria (both E and N gene results) for positivity of a clinical sample RT-LAMP was 98.5 per cent sensitive and 100 per cent specific for the presence of SARS-CoV-2 RNA. The single gene or both gene positivity was considered as positive for SARS-CoV-2. On the other hand, the negativity of a sample was considered by both gene negative results.
It is possible to use a constant temperature heat block in place of the expensive thermal cyclers, as RT-LAMP reactions are incubated at a single constant temperature. The assay can be used to increase the diagnostic testing capacity of COVID-19. The only limitation of the colorimetric RT-LAMP assay may be that it is not possible to be multiplexed.
Acknowledgment
The authors would like to thank Dr Balram Bhargava, DG-ICMR, Secretary DHR, Prof Priya Abraham, Director, National Institute of Virology, Pune, Dr Shailesh Pawar, OIC- NIV Mumbai Unit for moral help and support to carry out the experiments using clinical samples at NIV Mumbai Unit.
Financial support & sponsorship: The study was funded by the ICMR-NIV intramural grant.
Conflicts of Interest: None.
References
- Indian Council of Medical Research. ICMR SARS-CoV-2 (COVID-19) Testing Status 2021. Available from: https://www.icmr.gov.in/
- Indian Council of Medical Research-National Institute of Virology (SOP) 2020. Standard Operating Procedure for Detection of 2019 novel coronavirus in suspected human cases by rRT-PCR. Available from: https://www.icmr.gov.in/pdf/covid/labs/2_SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf
- Multicenter evaluation of the Cepheid xpert xpress SARS-CoV-2 test. J Clin Microbiol. 2020;58:e00926-20.
- [Google Scholar]
- Performance evaluation of Truenat™Beta CoV&Truenat™SARS-CoV-2 point-of-care assays for coronavirus disease 2019. Indian J Med Res. 2020;153:144-50.
- [Google Scholar]
- Rapid, accurate, nucleobase detection using FnCas9. medRxiv 2020 DOI:10.1101/2020.09.13.20193581
- [Google Scholar]
- Diagnosing COVID-19:The disease and tools for detection. ACS Nano. 2020;14:3822-35.
- [Google Scholar]
- Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10:97-101.
- [Google Scholar]
- A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12:eabc7075.
- [Google Scholar]
- 2020. Instructions for LabGun™ COVID-19 RT-PCR Kit. Available from: https://www.fda.gov/media/137483/download
- New England Biolabs. Bst 2.0 polymerase. Available from: https://international.neb.com/products/m0538-bst- 20-warmstart-dna-polymerase#Product%20Information
- New England Biolabs. WarmStart RTx reverse transcriptase. Available from: https://international.neb.com/products/m0380-warmstart-rtx-reverse-transcriptase#Product%20Information
- Eiken Genome Site. RT LAMP principle. Available from: http://loopamp.eiken.co.jp/e/lamp/rt_principle.html
- Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS One. 2020;15:e0238612.
- [Google Scholar]
- GLAPD:Whole genome based LAMP primer design for a set of target genomes. Front Microbiol. 2019;10:2860.
- [Google Scholar]
- Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci Rep. 2019;9:7400.
- [Google Scholar]






