Translate this page into:
Validation of an indigenous assay for rapid molecular detection of rifampicin resistance in presumptive multidrug-resistant pulmonary tuberculosis patients
ICMR's Task Force Expert Committee on TB Diagnostics: V.M. Katoch (Chairman), Anshu Prakash, Urvashi B. Singh, Rohit Sarin, Sarman Singh, Bindu Dey, Vijay Chowdhary, K.S. Sachdeva, S.D. Khaparde, Srikanth Tripathy, Camilla Rodrigues, Soumya Swaminathan
Participating Sites and Investigators: Rohit Sarin, Urvashi B. Singh, N.S. Gomathi, T. Kannan, Srikanth Tripathy, D.S. Chauhan, U.D. Gupta, V.P. Myneedu, Manpreet Bhalla, Anant Mohan, Anuj Bhatnagar
For correspondence: Dr Urvashi B. Singh, Department of Microbiology, All India Institute of Medical Sciences, Ansari Nagar, New Delhi 110 029, India e-mail: drurvashi@gmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
There is a need for an affordable, easy, high-sensitivity test usable at the peripheral health facility for diagnosis of drug-resistant (DR) tuberculosis (TB) to interrupt disease transmission. Nucleic acid amplification tests (NAATs) for early detection of DR-TB are ideal to bring testing near to the patient. Truenat™ MTB (Mycobacterium tuberculosis) and Truenat™ MTB-RIF (rifampicin) is an indigenous chip-based real-time polymerase chain reaction (PCR) based test for detection of multidrug-resistant (MDR) TB. The test involves extraction of DNA using automated, battery operated Trueprep instrument and real-time PCR performed on the Truelab analyzer. We report here multicentric validation of Truenat MTB-RIF for detection of DR-TB in suspected DR-TB patients.
Methods:
Consecutive patients aged 18-65 yr, with symptoms suggestive of TB and with a history of previous treatment, reporting to the National TB Elimination Programme (NTEP) clinics under four national institutes, namely AIIMS (All India Institute of Medical Sciences, New Delhi), NITRD (National Institute of Tuberculosis and Respiratory Diseases, New Delhi), NIRT (National Institute for Research in Tuberculosis, Chennai) and ICMR-National JALMA Institute for Leprosy and other Mycobacterial Diseases, Agra, were included in the study. Two sputum samples (one spot and one morning) were collected from each patient, after obtaining informed written consent. The samples were subjected to smear, GeneXpert and MGIT 960 culture (and drug susceptibility testing to RIF) (surrogate for MDR-TB) to serve as reference tests. The samples were coded to ensure blinding and subjected to Truenat MTB-RIF. Truenat MTB-RIF Version 1.5 was used for testing 1084 samples for RIF resistance, while Version 2.0 was used to test another 1201 samples.
Results:
Truenat MTB-RIF Version 1.5 in comparison with comprehensive laboratory reference standards yielded sensitivity and specificity of 76.2 and 94.7 per cent, respectively for the detection of RIF resistance in 1084 samples, collected across four sites. Based on the analysis of discordant samples, Version 2.0 of Truenat was developed by the manufacturer and this was further tested on additional 1201 samples, yielding a sensitivity of 87.5 per cent and specificity of 99.5 per cent.
Interpretation & conclusions:
Multicentric trial of Truenat™ MTB-RIF demonstrated a great potential of this point of care NAAT for detection of MDR-TB. The test would be useful in limited resource settings and inaccessible areas without need for any additional infrastructure.
Keywords
Drug resistant
tuberculosis
early diagnosis
nucleic acid amplification tests
Truenat MTB-RIF
tuberculosis
Early diagnosis of drug resistance is crucial for effective patient management and prevention of spread of severe forms of disease in India with about 64,000 multidrug-resistant-tuberculosis (MDR-TB) patients, equivalent to one-fifth of global MDR-TB population12. In order to design optimal regimen for treating such patients, liquid culture and drug susceptibility testing (DST) have been introduced in recent years with a shorter turn-around time but are expensive and need biosafety facilities. Molecular tests such as nucleic acid amplification tests (NAATs) with a shorter turn-around time, which can be performed directly on specimens, can accurately detect gene mutations responsible for drug resistance within hours. The WHO has endorsed Line Probe Assay (GenoType MTBDRplus, Hain Lifescience, Germany) and Xpert MTB/RIF (Mycobacterium tuberculosis/rifampicin) (Cepheid Inc., USA)1345. Xpert MTB/RIF is based on nested real-time polymerase chain reaction (PCR) and molecular beacon technology and is rapid, with a turn-around time of two hours6. It requires minimal biosafety facilities and is not prone to cross contaminations. The system is reported to have detection sensitivity higher than that of smear microscopy or solid culture7. Line probe assay (LPA) on the other hand takes 24-48 h for completion of the assay. Sensitive culture-based tests such as MGIT (mycobacteria growth indicator tube) and molecular tests such as LPA or GeneXpert MTB/RIF678 for TB are available under National TB Elimination Programme (NTEP)9101112. However, these technologies are expensive and manufactured outside the country, hence need for a point of care (POC) tool, which is affordable, sensitive and simple cannot be overstated. Truelab™ platform (Molbio Diagnostics, Goa) is one such tool, which can be effectively implemented in resource-limited settings not only for Mycobacterium tuberculosis (MTB) but also for detecting various other infections. Additional feature of this platform is that it is battery operated and portable and hence can be a true POC. In a pilot study13, it was reported that the test was able to detect TB rapidly with good sensitivity in comparison with a composite laboratory reference standard (CLRS). We report here the outcome of a multicentric study (ICMR Task Force on TB Diagnostics) designed to validate this platform in comparison to CLRS for detection of DR-TB in presumptive MDR-TB patients.
Material & Methods
Study design: The study was a double-blind, cross-sectional study to evaluate the diagnostic potential of Truenat MTB-RIF for diagnosis in comparison to conventional tests e.g. sputum smear, culture and GeneXpert MTB/RIF to diagnose MDR-TB. The study was designed to have real-time blinding and codes were provided by a statistician based at the National Institute for Research in Tuberculosis (NIRT), Chennai, India, the same day as the sample was received. After coding, the samples were sent for testing. The results were decoded and collated at the end of the study. The study was initiated after approval from the respective Institutional Ethics Committees and carried across four national institutes namely, All India Institute of Medical Sciences (AIIMS), New Delhi, National Institute of TB and Respiratory Diseases, New Delhi, NIRT, Chennai, and ICMR-National JALMA Institute for Leprosy & Other Mycobacterial Diseases, Agra. Written and informed consent was obtained from each participant prior to sputum collection.
Quality control: All four sites participated in inter-laboratory quality assurance for all methods. All sites functioned uniformly with high quality data and documentation. Samples were coded centrally and communicated to investigators at each site. Coded samples were subjected to the Candidate test while the reference tests were performed on the samples before coding.
Inclusion and exclusion criteria: The study was initiated in December 2014 and concluded in January 2016 at all four sites. All consecutive adults aged 18-65 yr with presumptive drug-resistant pulmonary TB patients, attending NTEP clinics were included in the study. Necessary details of the patients such as age, sex, address, contact details, prior history of TB treatment were recorded as per the NTEP, in the TB register. Exclusion criteria were inability of patient to produce two sputum samples of more than 4 ml, patients receiving anti-TB medication in the 60 days prior to testing and TB treatment started more than 48 h before sampling.Sample collection, sputum smear microscopy, MGIT 960 culture and DST, GeneXpert MTB/RIF and Truenat MTB-RIF were performed at respective institutions by trained staff.
Study population for evaluation of Truenat RIF Version 1.5: A total of 2586 presumptive MDR-TB patients met the inclusion criteria. Sputum specimens from the presumptive pulmonary MDR-TB patients under NTEP treatment were included in the study. The patient samples were collected individually on two consecutive days and two suptum samples were collected i.e. one on the spot and one in the next morning. The standard diagnostic tests i.e. sputum smear, culture and DST, GeneXpert MTB/RIF and Truenat MTB-RIF were done.
Sample size calculations: The sample size was calculated based on the following assumptions. The level of disagreement between the standard BACTEC MGIT 960 and the new method was estimated to be about 10 per cent with confidence interval of 95 per cent and interval width of five per cent. Based on the above assumptions, the minimum sample size was estimated to be 560 for each site with 20 per cent addition for possible dropouts or refusal.
Samples were delinked from patient details and assigned coded laboratory numbers. Following blinding, smear, GeneXpert MTB/RIF and Truenat MTB-RIF assays were performed directly on the sample. In the case of Truenat, DNA was extracted from the sample and subjected to MTB detection and if positive for MTB, testing for RIF resistance was done as an add-on test. The samples were pooled for conducting the molecular tests, GeneXpert MTB/RIF and LPA. Pooled samples were coded by the statistician at NIRT, Chennai, before subjecting to Truenat tests.
The remaining portion of the sputum sample was decontaminated by the standard N-acetyl-L-cysteine-sodium hydroxide method and resultant pellet was used for inoculation of MGIT960, and for LPA. Confirmed MTB positive cultures were tested for susceptibility to RIF by MGIT 960. All tests were performed as per manufacturers' instructions or standard approved laboratory protocols.
-
(i)
AFB smears: The processed specimens were used for making smears for all samples. All the smears were stained by the Ziehl-Neelsen method and examined with a light microscope14.
-
(ii)
Liquid culture (MGIT 960): The decontaminated samples (0.5 ml) were inoculated into MGIT 960 non-radiometric automated isolation system (Becton Dickinson, Sparks, MD, USA), Positive cultures were confirmed using smear microscopy and TBc Identification test (TBcID, Becton Dickinson, Sparks, USA) as MTB15.
-
(iii)
Liquid culture (DST): DST for RIF was performed with the MGIT 960 system, using the WHO recommended standard critical concentration of 1 μg/ml RIF and 0.1 μg/ml isoniazid (INH). Standard protocol was followed according to the manufacturer's instructions15.
-
(iv)
LPA: Decontaminated samples were used for the test and pellets were re-suspended in 1.5 ml of phosphate buffer. Assay was performed as per the manufacturer's instructions16.
-
(v)
Xpert MTB-RIF: The assay was performed as per the manufacturer's instructions17.
-
(vi)
Truenat MTB and Truenat MTB/RIF: The Truenat™ MTB is a chip-based NAAT for detection of MTB from sputum samples. The test involves sputum processing using Trueprep™ MAG (a nano-particle-based protocol run on a battery operated device) and real-time PCR performed on the Truelab™ Uno analyzer (hand-held battery operated thermal cycler). Sputum samples were processed as per the manufacturer's instruction on Trueprep-MAG sputum kit with a starting volume of 500 μl being added to the sample pre-treatment tube. A total of 5 μl DNA was extracted and added to Truenat MTB microchip containing lyophilized master-mix and real-time PCR was performed using a pre-programmed profile set on the device. Master-mix was lyophilized and contained primers and a probe specific to MTB. A total of 4 μl of extracted DNA was mixed with 6 μl of Truenat master-mix. The cycles of real-time PCR were as follows: one minute at 95°C, and 45 cycles of 10 sec at 95°C and 34 sec at 58°C18.
Sequencing of discordant samples: Samples from AIIMS site giving discordant results using the two systems, GeneXpert and Truenat were sequenced. The rpoB gene was amplified using previously published primers from bacterial DNA67. Cycle sequencing was performed using Big-Dye Terminator Ready Reaction Cycle Sequencing Kit (PE Biosystems, USA) and the products loaded in the ABI Prism 310 Genetic Analyzer (PE Biosystems). DNA sequences thus obtained were aligned for homology using Basic Local Alignment Search Tool algorithm in the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and analyzed for mutations with Genedoc Multiple Sequence Alignment Editor (http://www.psc.edu/biomed/genedoc).
Study population for evaluation of modified version (Truenat RIF Version 2.0): Truenat Version 1.5 did not have control probe for melting temperature (Tm) correction (to normalize the Tm values from the same chip, Probe melt Tm deviates due to many reasons including the elute quality and presence of ions), hence samples which fell in the Tm cut-off range ±0.5°C were labelled as indeterminate. The manufacturers incorporated changes to include a control probe and came out with Version 2.0. This retrospective study on Version 2.0 was done at NIRT, Chennai, and was approved by the institutional ethics committee. Left over de-identified sputum samples from 1201 consecutive presumptive MDR-TB patients attending NTEP clinics of Chennai and Kanchipuram districts, Tamil Nadu, India, that were stored in deep freezer (−80°C) were included in the study. After excluding samples that yielded invalid, indeterminate and contaminated culture results, data from 1075 samples were analyzed.
Statistical analysis: Sensitivity, specificity, positive predictive values (PPVs) and negative predictive values (NPVs) were calculated using STATA version 16 (StataCorp LP, College Station, TX, USA). Data analysis was done using STATA 13.1, College Station, Texas 77845 USA. Sensitivity, specificity, positive predictive value, negative predictive value, likelihood ratio positive (LR+) and likelihood ratio negative (LR−) were derived at 95 per cent level of significance.
Results
This study was aimed to evaluate the Truenat MTB-RIF (Version 1.5) for detection of resistance to RIF in comparison with the reference standards BACTEC MGIT960, GeneXpert MTB/RIF and LPA in terms of sensitivity, specificity, efficiency, predictive values and time for detection. Performance of Truenat MTB-RIF for detection of RIF resistance was determined by comparing the results with comprehensive laboratory reference standard (Tables I and II).
| Culture | Xpert | Line probe assay | Truenat=1184 | ||
|---|---|---|---|---|---|
| Resistant (R) (%) | Susceptible (S) (%) | Total | |||
| R | R | R | 138 (76.2) | 43 (23.8) | 181 |
| R | R | S | 13 (52) | 12 (48) | 25 |
| R | S | R | 2 (22.2) | 7 (77.8) | 9 |
| R | S | S | 3 (5.6) | 51 (94.4) | 54 |
| S | R | R | 14 (58.3) | 10 (41.7) | 24 |
| S | R | S | 23 (54.8) | 19 (45.2) | 42 |
| S | S | R | 9 (52.9) | 8 (47.1) | 17 |
| S | S | S | 44 (5.3) | 788 (94.7) | 832 |
Truenat MTB/RIF assay in comparison with the pooled cumulative gold standard (rows 1 and 8) yielded sensitivity and specificity of 76.2 and 94.7 per cent, respectively for detection of RIF resistance. Among 54 specimens where both the gold standard molecular tests yielded a susceptible pattern (row 4) Truenat exhibited an agreement of 94.4 per cent. Among 24 specimens where the gold standard molecular tests yielded a resistance pattern (row 5) Truenat exhibited an agreement of 58.3 per cent
| Candidate test | Comprehensive laboratory reference standard | |||||
|---|---|---|---|---|---|---|
| R | S | Total | ||||
| Sensitivity | 0.76 (0.69, 0.82) | |||||
| Truenat | R | 138 | 44 | 182 | Specificity | 0.95 (0.93, 0.96) |
| S | 43 | 788 | 831 | Positive predictive value | 0.76 (0.69, 0.82) | |
| Total | 181 | 832 | 1013 | Negative predictive value | 0.95 (0.93, 0.96) | |
| Positive likelihood ratio | 14.42 (10.69, 19.44) | |||||
| Negative likelihood ratio | 0.25 (0.19, 0.33) | |||||
Truenat in comparison with the cumulative gold standard yielded a sensitivity, specificity, positive predictive value, negative predictive value are 76.2, 94.7, 75.8 and 94.7 per cent, respectively for detection of RIF resistance. R, resistant; S, susceptible
Candidate molecular test Truenat MTB-RIF was found to detect Rif resistance in 138 (76.2%) of 181 samples that were resistant by the comprehensive laboratory reference standard. The observed agreement was 91.4 per cent and the kappa value was 0.708 with good strength of agreement. In addition, Truenat could rule out RIF resistance in 94.7 per cent samples, when compared to comprehensive laboratory reference standards.
There were five samples, which showed discordance in RIF resistance detection between Truenat and Xpert, at AIIMS centre. These cultures were subjected to target sequencing for rpoB gene. Three strains with mutation at position 518 of rpoB gene (traditional numbering system), were picked by Truenat, while GeneXpert missed it and in two cultures S531L mutation in the rpoB gene was missed by Truenat (Table III).
| Truenat MTB-RIF result | GeneXpert MTB/RIF result | Reported MIC μg/ml19 | Sanger sequencing rpoB gene mutation (Escherichia coli numbering system) | Treatment given (based on) | GenBank accession number |
|---|---|---|---|---|---|
| Mtb detected; susceptible | Mtb detected; resistant | 32 | Serine 531 leucine | MDR (GeneXpert result) | MT437290 |
| Mtb detected; susceptible | Mtb detected; resistant | 32 | Serine 531 leucine | MDR (GeneXpert result) | MT437291 |
| Mtb detected; resistant | Mtb detected; susceptible | 32 | 518 deletion of asparagine | MDR (clinical presentation) | MT437292 |
| Mtb detected; resistant | Mtb detected; susceptible | 32 | 518 deletion of asparagine | MDR (clinical presentation) | MT437293 |
| Mtb detected; resistant | Mtb detected; susceptible | 32 | 518 deletion of asparagine | MDR (clinical presentation) | MT437294 |
Both the mutations reported here lead to clinical RIF resistance (>1 μg/ml). MDR, multidrug-resistant; Mtb, Mycobacterium tuberculosis; MIC, minimum inhibitory concentration
The discordant samples (Table II) detected as RIF resistant using laboratory reference standards but sensitive by Truenat MTB-RIF (n=43) and those sensitive by using laboratory reference standards but resistant by Truenat MTB-RIF (n=44) were analyzed carefully. With the repeat analysis, 22 of 44 and 6 of 43 samples were classified as indeterminates.
These results were instrumental in guidance for improvement in the Truenat system. The upgraded device included three wavelengths instead of two in the older device, to normalize the data. An internal control probe was designed and included in the same chip. Any shift of the Tm value of IC probe was used to normalize the observed Tm values of Probe 1 and Probe 2 (Figure).
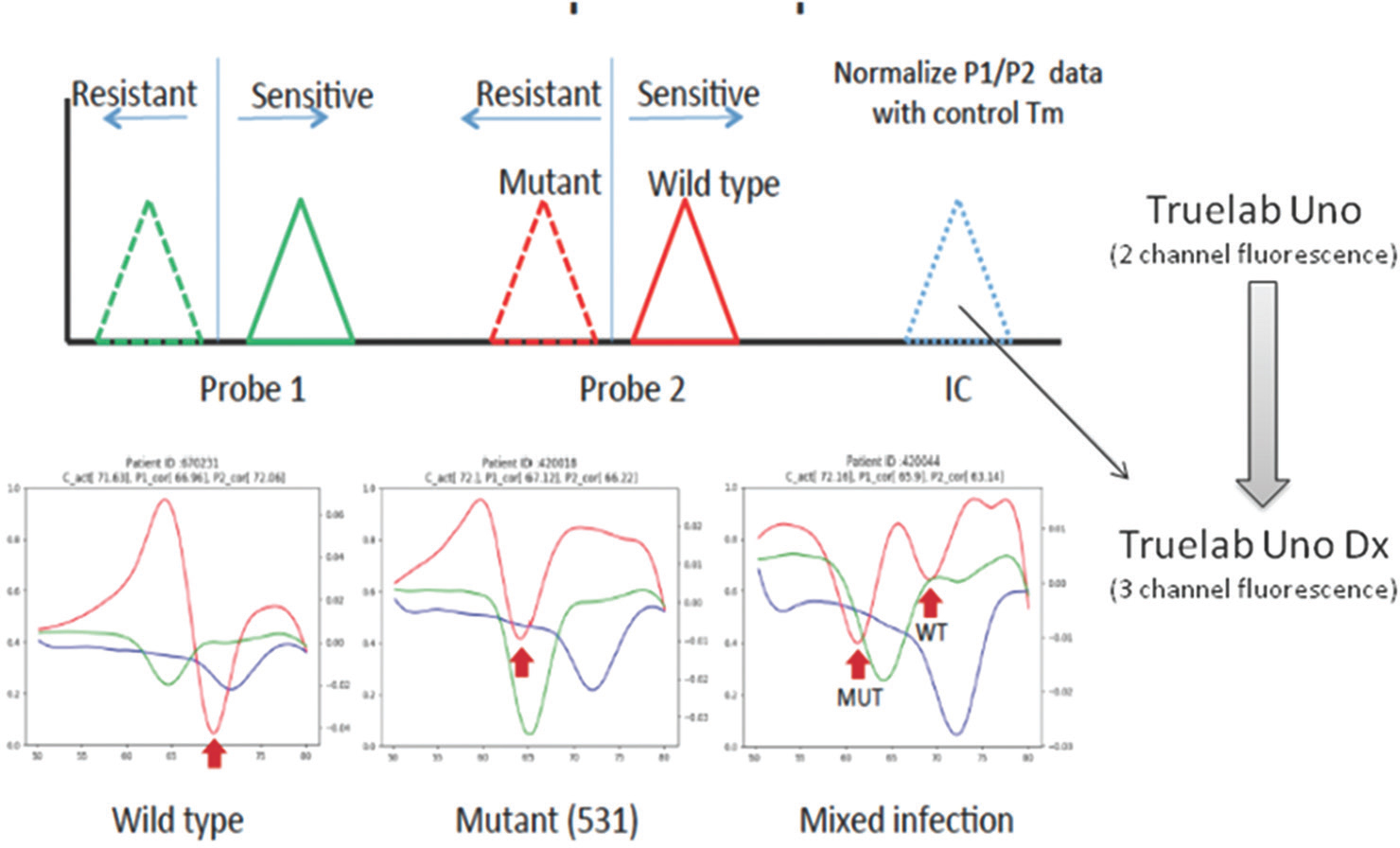
- Differences in Truenat Versions 1.5 and 2.0: Inclusion of internal control and third wavelength in Version 2.0 enabled better distinction between rifampicin sensitive and resistant Mycobacterium tuberculosis.
Algorithm modification for Truenat RIF Version 1.5: Initial analysis was based on the probe Tm cut-off values generated using a small, characterized sample panel to distinguish wild type and mutant strains. An algorithm correction was applied to call samples that fall within ±0.5°C around the nominal cut-off Tm as indeterminate. With this it was observed that the Truenat MTB-RIF assay sensitivity and specificity were significantly improved. Diagnostic efficacy of Truenat MTB-RIF was re-assessed after removing indeterminate samples. Sensitivity increased to 78.53 per cent and specificity became 97.37 per cent. Data also showed PPV of 86.49 per cent and NPV of 95.48 per cent (Figure).
Evaluation of modified version (Truenat RIF Version 2.0): The modified device was subjected to further evaluation in another set of samples. The study included 1201 samples of which 906 were smear positive and 295 were smear negative. After excluding samples with invalid, indeterminate or contaminated results, analysis was done for 1075 samples. Interpretable results from all DSTs were available for 633 samples and were analyzed for overall performance (Table IV).
| MGIT | Xpert | LPA | Truenat | Total | |
|---|---|---|---|---|---|
| Resistant (R) (%) | Susceptible (S) (%) | ||||
| R | R | R | 28 (87.5) | 4 (12.5) | 32 |
| R | R | S | 5 (100) | 0 (0) | 5 |
| R | S | R | 0 (0) | 2 (100) | 2 |
| R | S | S | 0 (0) | 21 (100) | 21 |
| S | R | R | 11 (73.3) | 4 (26.7) | 15 |
| S | R | S | 4 (80) | 1 (20) | 5 |
| S | S | R | 0 | 0 | 0 |
| S | S | S | 3 (0.5) | 550 (99.5) | 553 |
| Total | 51 (8.1) | 582 (91.9) | 633 | ||
Truenat MTB/RIF assay in comparison with the comprehensive laboratory reference standards (rows 1 and 8) yielded a sensitivity and specificity of 87.5 and 99.5 per cent, respectively for detection of RIF resistance. MGIT, mycobacteria growth indicator tube; LPA, line probe assay
Specificity of TB detection by Truenat: In order to study the specificity of TB detection, non-TB disease controls were enrolled from Pulmonology clinic at AIIMS, New Delhi. One hundred and fifty patients with proven aetiology/diagnosis other than TB, such as bronchiectasis, chronic obstructive pulmonary disease, asthma, lung carcinoma and pneumothorax were enrolled and subjected both to reference tests and Truenat test. The laboratory reference tests were negative for all these patients. Truenat test also turned out negative for all the patients, giving a specificity of 100 per cent.
Discussion
Early diagnosis of diseases transmitted through droplets or airborne route, is important for control of disease spread in the community. Difficult to treat MDR-TB is one such disease. Newer technologies have brought a sea change in this field, obviating current empirical medical practice, however, diagnosis in difficult to reach and remote settings remains a challenge. The WHO approved LPA and GeneXpert MTB/RIF are available at the district level in India, under NTEP, while Truenat system from Molbio Diagnostics brought PCR technology as a POC test, which could be used at the District Microscopy centres. The added advantage of battery operated devices and storage of consumables at ambient temperature are features that prepare it for use in a less equipped laboratory.
The diagnosis of such ailments needs to be carefully followed by optimum treatment. Hence, the communication of diagnosis of cases of MDR-TB to the treating physician becomes important for early initiation of treatment. Truelab devices are network data enabled (through Wifi, Bluetooth and GSM modes) and transfer data to cloud server for centralized monitoring and surveillance. Real-time access to diagnostic data, independent of manual reporting, is crucial to implement, monitor and ensure large healthcare programmes like TB control.
This study was conducted in two phases. Phase I evaluated detection of TB from presumptive TB patients and Phase II evaluated RIF resistance detection in comparison with standard tests. In Phase I study, overall detection for Truenat MTB was higher as compared to GeneXpert MTB/RIF20. Overall performance of Truenat MTB-RIF was observed with sensitivity of 76.2 per cent and specificity of 98.4 per cent. On analyzing probable causes of discordant results, it was found that adding an internal control to normalize the probe melt Tm in the Truenat MTB-RIF assay could improve accuracy of detection. Hence, it was suggested to upgrade the device hardware and software by incorporating additional fluorescence channels. Subsequent laboratory evaluation gave promising results, with RIF resistance detection sensitivity improving to 87.5 per cent and specificity to 99.5 per cent.
The device has a potential in detecting resistant cases in a limited resource setting and curtailing cost of transporting samples to district hospitals or treatment units (TUs) where GeneXpert machines are placed. Currently, testing of DR-TB cases is completely dependent on the GeneXpert centres, which are often located far from Designated Microscopy Center (DMC). Using Truenat as an additional diagnostic test may be help in diagnosing MDR-TB patients in remote areas. The indigenous test would also obviate the need for importing expensive consumables for equally expensive GeneXpert systems. In addition, the ease of service and support within the country along with a cloud based tracking of results from peripheral sites could enable the geo-tracking of DR patients on treatment under the NTEP.
Globally, TB control programmes would need to rely on fast and accurate detection of TB and MDR-TB at the community level with a point-of-care test, especially in a high burden country such as India. The introduction of Truenat can serve as an acceptable and practical solution to detect missing millions, which may be going undetected due to inaccessibility and operational hurdles. Although more studies are required to throw light on this diagnostic test, the present evidence appears promising, to take this technology forward.
Acknowledgment:
Authors thank all the patients that made this study possible, and also thank the technical staff of TB laboratories in all four sites for their support.
Financial support & sponsorship: This study was supported by the Department of Biotechnology, Government of India. Machines and assay chips were provided by the Molbio Diagnostics, Goa.
Conflicts of Interest: None.
References
- Global Tuberculosis Report 2019. Geneva: WHO; 2019.
- WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment 2019
- New and improved tuberculosis diagnostics: Evidence, policy, practice, and impact. Curr Opin Pulm Med. 2010;16:271-84.
- [Google Scholar]
- Framework for Implementing New Tuberculosis Diagnostics 2010
- Present position of microscopy and of culture in diagnostic mycobacteriology. Zentralbl Bakteriol Mikrobiol Hyg A. 1985;260:81-7.
- [Google Scholar]
- Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005-15.
- [Google Scholar]
- Evaluation of the analytical performance of the Xpert MTB RIF assay. J Clin Microbiol. 2010;48:2495-501.
- [Google Scholar]
- Evaluation of the cepheid Xpert MTB RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Cli Microbiol. 2011;49:1621-3.
- [Google Scholar]
- Advances in tuberculosis diagnostics: The Xpert MTB RIF assay and future prospects for a point-of-care test. Lancet Infect Dis. 2013;13:349-61.
- [Google Scholar]
- Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB RIF test for diagnosis of tuberculosis and multidrug resistance: A multicentre implementation study. Lancet. 2011;377:1495-505.
- [Google Scholar]
- Xpert® MTB RIF for point-of-care diagnosis of TB in high-HIV burden, resource-limited countries: Hype or hope? Exp Rev Mol Diagn. 2010;10:937-46.
- [Google Scholar]
- Achieving STOP TB Partnership goals: Perspectives on development of new diagnostics, drugs and vaccines for tuberculosis. Trop Med Int Health. 2011;16:819-27.
- [Google Scholar]
- Rapid diagnosis of Mycobacterium tuberculosis with Truenat MTB: A near-care approach. PLoS One. 2013;8:e51121.
- [Google Scholar]
- Laboratory Series in Tuberculosis Control Part II, Microscopy WHO/TB/98258. Geneva, Switzerland: WHO; 1998.
- MGIT Procedure Manual. Geneva, Switzerland: Foundation for Innovative New Diagnostics (FIND); 2006.
- [Google Scholar]
- 2012. GenoLyse® Hain VER 1.0. Instructions for Use, IFU-51610-09. Germany: Hain Lifescience GmbH; Available from: http://www.hain-lifescience.de/en/instructions-for-use.html
- Truenat™ MTB, Manufacturer's Guidelines. Goa: Molbio Diagnostics Pvt. Ltd; 2017.
- Mutation analysis of mycobacterial rpoB genes and rifampin resistance using recombinant Mycobacterium smegmatis. Antimicrob Agents Chemother. 2012;56:2008-13.
- [Google Scholar]
- Multicentric validation of indigenous molecular test Truenat™ MTB for detection of Mycobacterium tuberculosis in sputum samples from presumptive pulmonary tuberculosis patients in comparison with reference standards. Indian J Med Res. 2020;152:378-85.
- [Google Scholar]






