Translate this page into:
Mapping the genomic landscape & diversity of COVID-19 based on >3950 clinical isolates of SARS-CoV-2: Likely origin & transmission dynamics of isolates sequenced in India
*For correspondence: seyedhasnain@gmail.com
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Sir,
The COVID-19 pandemic has stalled the world and catapulted the global health systems into unprecedented chaos. More than 200 countries have been affected by this pandemic, resulting in 2.54 million cases in a short period of time and >0.17 million deaths (as of April 23, 2020), with a mere 0.7 million recoveries1. The movement of COVID-19 hotspot from China to Europe, and now to the USA, has been partly due to the staggered restrictions in global travel and partly due to potent transmission through asymptomatic carriers2.
India, with 21,393 cases and 681 deaths (as of April 23, 2020)1, had the lowest figures for any country of the comparable population (0.5 deaths per million population). International travellers or their close contacts formed the majority of initially reported cases. The delayed onset of COVID-19 in India has given it an edge, which allowed it to impose severe restrictions to contain the local spread 345.
In our in-depth analyses of 1500+ genomes, variability among clinical isolates was shown along the timeline, leading to distinct clustering of SARS-CoV-2 across the globe (unpublished observation). It was predicted, based on the aggregation propensity of the spike protein in the Wuhan and other isolates of SARS-CoV-2, that this virus would exhibit very high transmissibility and confer survival fitness67. Genetic diversity of the virus increases with disease progression and can be utilized to model the evolution and propagation of the disease6 Recently, phylogenetic network analysis of 160 SARS-CoV-2 genome samples showed a parallel evolution of the virus and its evolutionary selection in their human hosts8. Similar whole-genome analyses of the Indian isolates and their comparison with global isolates can provide a better understanding of dominant clades within the population and unveil targets for developing specific interventions.
In the present study, machine learning-based t-SNE analysis of global clinical isolates has been utilized to segregate the clinical isolates into clusters while accommodating the outliers910. Whole-genome analysis of 3968 global isolates obtained from GISAID (Global initiative on sharing all influenza data)11, including 25 SARS-CoV-2 genomes sequenced in India [next-genome sequencing (NGS) data submitted by the ICMR-National Institute of Virology, Pune, India] and presented in (Figure 1) (
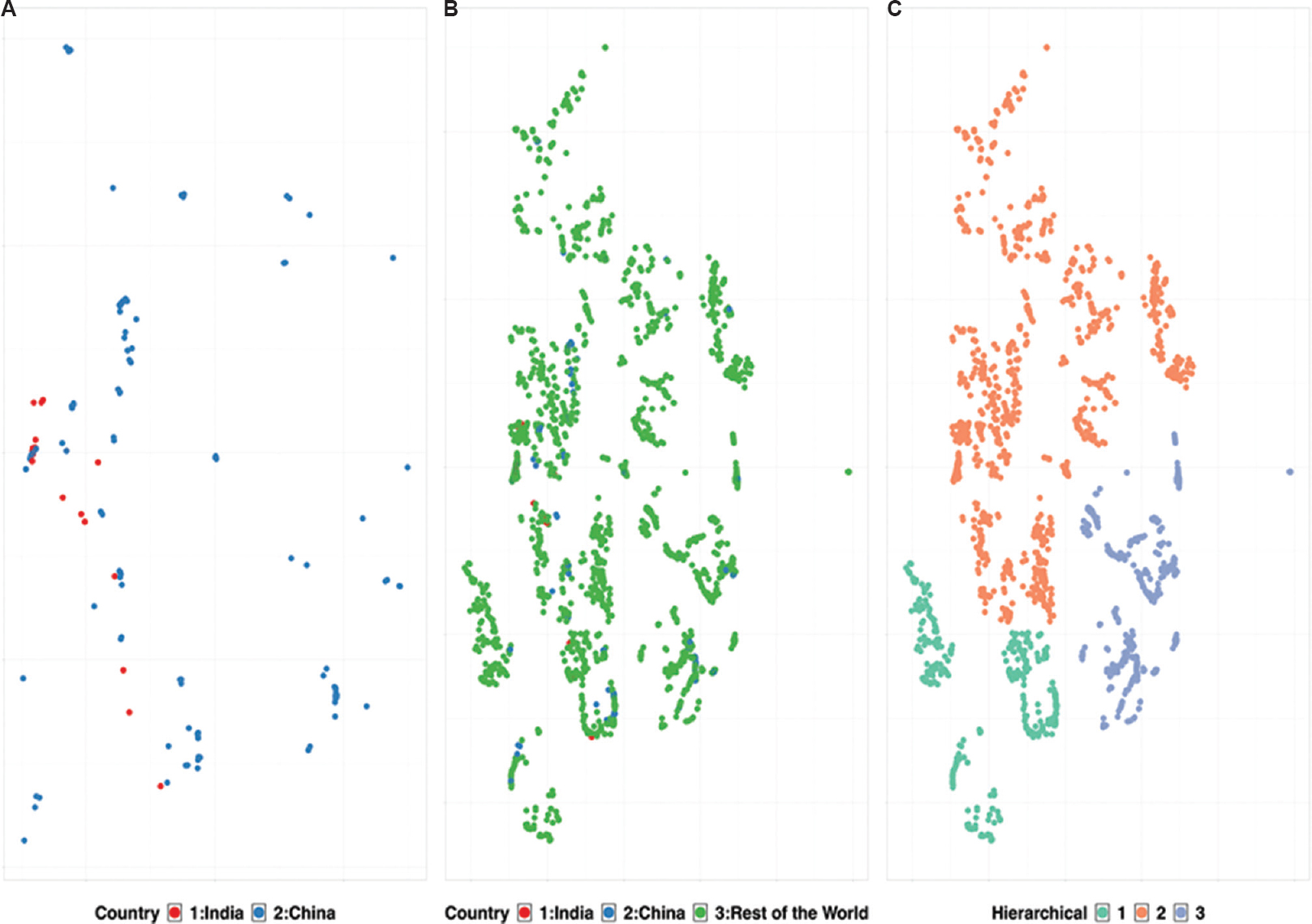
- Whole-genome-based t-SNE clustering of 3968 clinical isolates. (A) Comparative genome-based clustering of Indian isolates (red) with Chinease isolates (blue). (B) Comapartive genome-based clustering of Indian isolates (red) and Chinese isolates (blue) with rest of the world (green). (C) Diversity in clinical isolates showing three distinct clustering using hierarchical clustering on the t-SNE clusters. (t-SNE: https://github.com/jdonaldson/rtsne).
| Accession ID | Contact History | Gender | Age | Virus name | Location | Collection date | Hierarchical Cluster ID |
|---|---|---|---|---|---|---|---|
| EPI_ISL_413523 | Travel history to China | Male | 23 | hCoV-19/India/1-31/2020 | Asia/India/Kerala | 2020-1-31 | 2 |
| EPI_ISL_420543 | Italian tourist | Female | 73 | hCoV-19/India/763/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420544 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020763/2020 | Asia/India | 2020 | 1 |
| EPI_ISL_420545 | Italian tourist | Female | 77 | hCoV-19/India/770/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420546 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020770/2020 | Asia/India | 2020 | 1 |
| EPI_ISL_420547 | Italian tourist | Female | 70 | hCoV-19/India/772/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420548 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020772/2020 | Asia/India | 2020 | 2 |
| EPI_ISL_420549 | Italian tourist | Female | 65 | hCoV-19/India/773/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420550 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020773/2020 | Asia/India | 2020 | 2 |
| EPI_ISL_420551 | Indian contact of Italian tourist | Male | 59 | hCoV-19/India/777/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420552 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020777/2020 | Asia/India | 2020 | 2 |
| EPI_ISL_420553 | Italian tourist | Male | 66 | hCoV-19/India/781/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420554 | Vero CCL81 isolate P1 | NA | NA | hCoV-19/India/2020781/2020 | Asia/India | 2020 | 2 |
| EPI_ISL_420555 | Indian contact of Indian Patient having travel history to Italy | Female | 37 | hCoV-19/India/c32/2020 | Asia/India | 2020-3-3 | 2 |
| EPI_ISL_420556 | Vero CCL81 isolate P1 | NA | hCoV-19/India/2020c32/2020 | Asia/India | 2020 | 1 | |
| EPI_ISL_421662 | Indian citizen sampled at Iran | Male | 68 | hCoV-19/India/1073/2020 | Asia/India | 2020-3-10 | 2 |
| EPI_ISL_421663 | Indian citizen sampled at Iran | Male | 45 | hCoV-19/India/1093/2020 | Asia/India | 2020-3-10 | 2 |
| EPI_ISL_421664 | Indian citizen sampled at Iran | Male | 72 | hCoV-19/India/1100/2020 | Asia/India | 2020-3-10 | 2 |
| EPI_ISL_421665 | Indian citizen sampled at Iran | Male | 43 | hCoV-19/India/1104/2020 | Asia/India | 2020-3-10 | 1 |
| EPI_ISL_421666 | Indian citizen sampled at Iran | Female | 54 | hCoV-19/India/1111/2020 | Asia/India | 2020-3-10 | 2 |
| EPI_ISL_421667 | Indian citizen sampled at Iran | Male | 66 | hCoV-19/India/1115/2020 | Asia/India | 2020-3-10 | 2 |
| EPI_ISL_421669 | Indian citizen sampled at Iran | Female | 70 | hCoV-19/India/1616/2020 | Asia/India | 2020-3-12 | 2 |
| EPI_ISL_421670 | Indian citizen sampled at Iran | Female | 50 | hCoV-19/India/1617/2020 | Asia/India | 2020-3-12 | 2 |
| EPI_ISL_421671 | Indian citizen sampled at Iran | Female | 55 | hCoV-19/India/1621/2020 | Asia/India | 2020-3-12 | 2 |
| EPI_ISL_421672 | Indian citizen sampled at Iran | Male | 59 | hCoV-19/India/1644/2020 | Asia/India | 2020-3-12 | 2 |
The initial cases reported from India had a travel history to China, which explained its position in a Chinese cluster5 (Fig. 2). The travel ban from China to India, in early February 2020, has prevented the large-scale spill-over directly from China to the Indian Sub-continent. However, various isolates transmitted from other South-East Asian countries might fall in the same cluster. The overlap of Indian samples majorly with European samples (
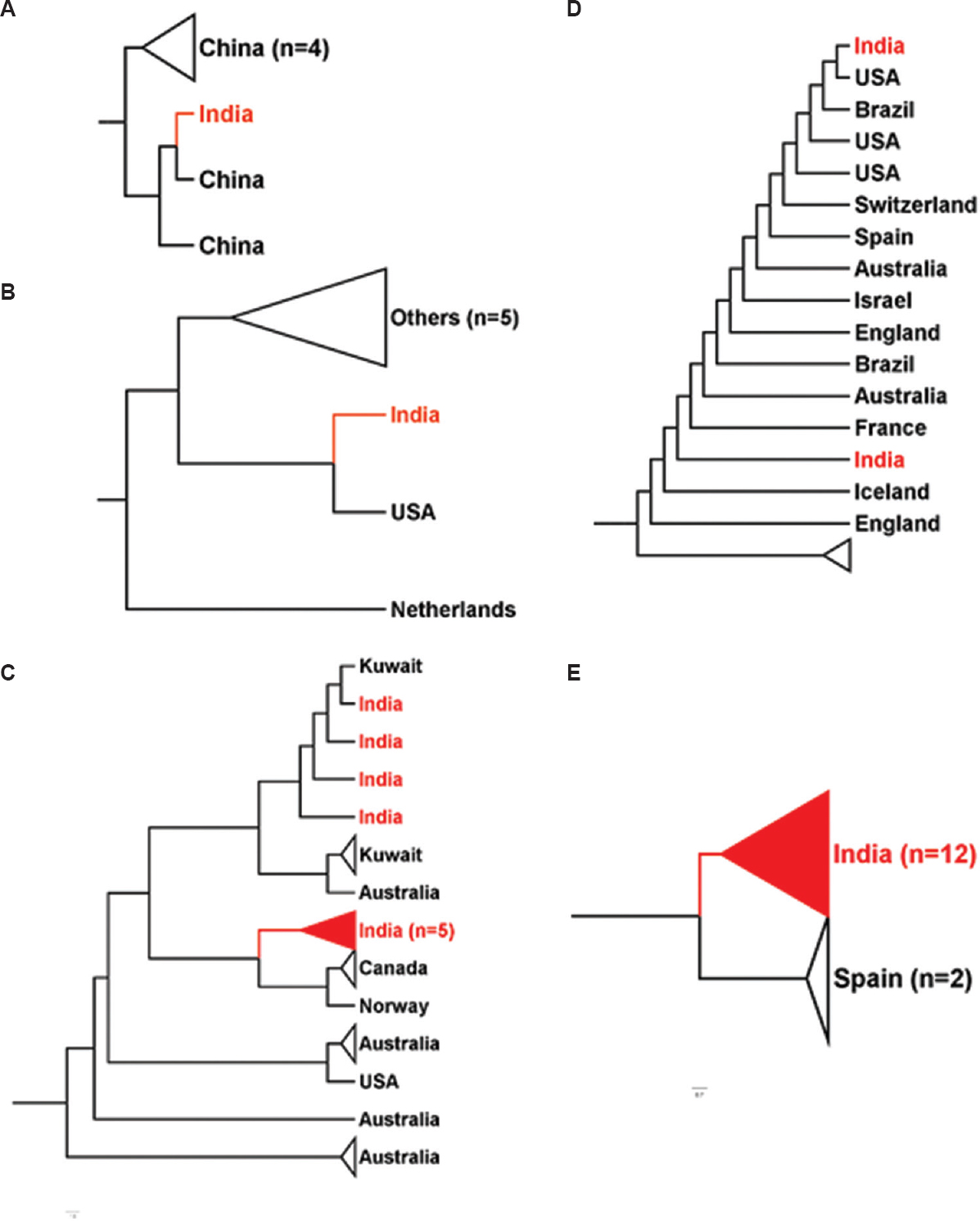
- Position of various Indian isolates with other nations. (A-E) Clustering of SARS-CoV-2 genome sequences from India (red) with other nations around the globe. Indian samples clustered with samples from different nations – China, Kuwait, Canada, USA and Spain in whole-genome-based clustering. Figures were generated using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Hierarchical-based clustering further yielded exciting outcomes on the inter-continent transmission of COVID-19. The segregation of SARS-CoV-2 genomes into three clades indicates the emergence of evolutionary diversity (Fig. 1C). The heterogeneity of these clusters, grouped along with Chinese counterparts, validates a global spill-over event originating from Wuhan512. Hierarchical cluster 2 in
The conservation of an amino acid in any protein sequence denotes its functional importance1314 as it undergoes fewer amino acid replacements or is more likely to substitute amino acids with similar biochemical properties. The amino acid conservation is inversely proportionate to the evolutionary rate. This is a valuable gauge of the evolutionary divergence and the analogous genomic regions. Sequence similarity between the open reading frames (ORFs) of Indian isolates and the initial sample collected in Wuhan unravels conservation in five ORFs corresponding to envelope protein, membrane glycoprotein, ORF6, ORF7b and ORF10 proteins (Fig. 3A). On the contrary, a number of mutations were observed in ORF1a, ORF1b, spike protein (surface glycoprotein), ORF3a, ORF7a, ORF8 and nucleocapsid phosphoprotein (
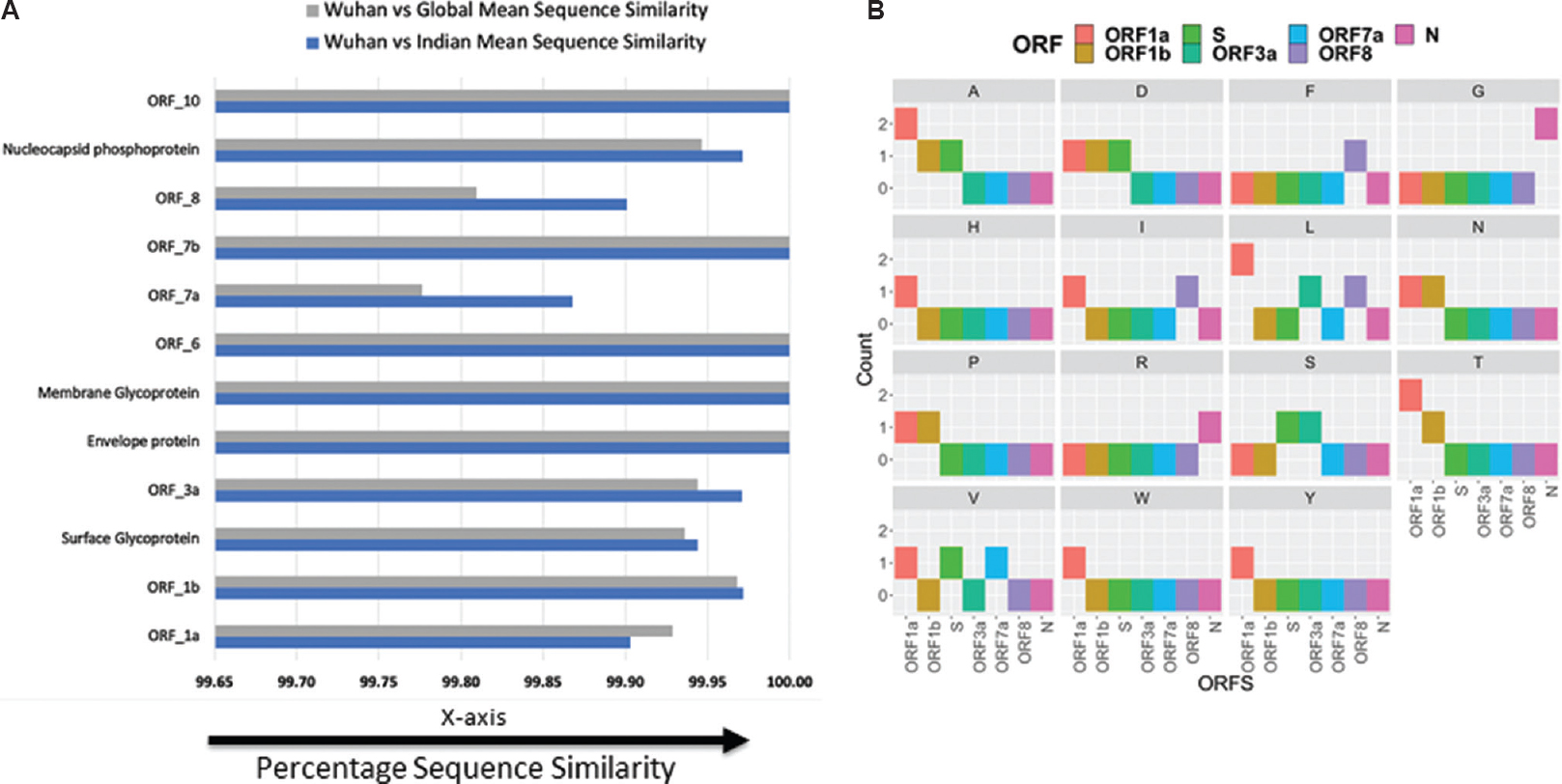
- Sequence similarity and mutation analysis of open reading frames. (A) Comparison of mean sequence similarity for open reading frames between Indian and global isolates with Wuhan strain. (B) Qualitative analysis on type of mutations occurring in non-conserved open reading frames (ORFs) of Indian isolates compared to Wuhan strain.
| ORF | Position | Amino acid in reference | Mutated amino acid | Mutation |
|---|---|---|---|---|
| ORF1a | 207 | Arginine (R) | Cysteine | R->C |
| ORF1a | 378 | Valine (V) | Isoleucine (I) | V->I |
| ORF1a | 1515 | Serine (S) | Phenylalanine (F) | S->F |
| ORF1a | 2796 | Methionine (M) | Isoleucine (I) | M->I |
| ORF1a | 3606 | Leucine (L) | Phenylalanine (F) | L->F |
| ORF1b | 314 | Proline (P) | Leucine (L) | P->L |
| S | 614 | Aspartic acid (D) | Glycine (G) | D->G |
| ORF7a | 74 | Valine (V) | Phenylalanine (F) | V->F |
| Column 1 ORF |
Column 2 Name |
Wuhan vs Indian | Wuhan vs Global | ||
|---|---|---|---|---|---|
| Indian Mean Similarity | Indian GMean Similarity | Global Mean Similarity | Global GMean Similarity | ||
| ORF5 | ORF_1a | 99.90 | 99.90 | 99.93 | 99.93 |
| ORF1 | ORF_1b | 99.97 | 99.97 | 99.97 | 99.97 |
| ORF2 | Surface Glycoprotein | 99.94 | 99.94 | 99.94 | 99.94 |
| ORF3 | ORF_3a | 99.97 | 99.97 | 99.94 | 99.94 |
| ORF4 | Envelope protein | 100.00 | 100.00 | 100.00 | 100.00 |
| ORF6 | Membrane Glycoprotein | 100.00 | 100.00 | 100.00 | 100.00 |
| ORF7 | ORF_6 | 100.00 | 100.00 | 100.00 | 100.00 |
| ORF8 | ORF_7a | 99.87 | 99.87 | 99.78 | 99.78 |
| ORF9 | ORF_7b | 100.00 | 100.00 | 100.00 | 100.00 |
| ORF10 | ORF_8 | 99.90 | 99.90 | 99.81 | 99.81 |
| ORF11 | Nucleocapsid phosphoprotein | 99.97 | 99.97 | 99.95 | 99.95 |
| ORF12 | ORF_10 | 100.00 | 100.00 | 100.00 | 100.00 |
Evolutionary divergence, corroborated by epidemiological data, is a valuable tool to implement appropriate measures against this pandemic. The population density of India and the presence of functionally distinct isolates in the Indian population raise concerns and warrant an urgent need for higher sampling rate for better assessment of the evolution of SARS-CoV-2 in India. The situation is further confounded by the fact that many of these Indian isolates submitted in databanks include those of Indians living in Iran, Italian tourists visiting India, and also contains samples cultured in vitro.
In conclusion, a whole-genome diversity analysis of 3968 global clinical isolates, including 25 isolates sequenced in India, of SARS-CoV-2 was done. The variations in different open reading frames (ORFs) of SARS-CoV-2, which drives the formation of distinct Indian clusters and functional heterogeneity, were highlighted. Five ORFs corresponding to envelope protein, membrane glycoprotein, ORF6, ORF7b and ORF10 were found to be highly conserved, while a number of mutations were observed in ORF1a, ORF1b, spike protein, ORF3a, ORF7a, ORF8 and nucleocapsid phosphoprotein. Generating diverse genomic datasets will provide insight into the propagation dynamics of COVID-19, leading to a better understanding of pathogenesis and evolution of SARS-CoV-2, which will eventually lead to better intervention methods.
SUPPLEMENTARY INFORMATION
Mapping the genomic landscape and diversity of COVID-19 based on >3950 clinical isolates of SARS-CoV-2: Likely origin and transmission dynamics of isolates sequenced in India
Supplementary Fig. 1
Supplementary Fig. 1 Position of Indian genome sequences in sub-cluster a, b, and c with respect to other global genome sequences. Panel I shows presence of Indian isolates in three distinct sub-clusters with respect to Chinese isolates and remaining global isolates. Panel II highlights the placement of Indian clusters in two of the three hierarchical clusters (a, b, and c) obtained from t-SNE whole genome clustering of 3968 sequences. Panel III displays the prevalence of samples from various continents. Continent codes; Africa (Green), Australia (Orange), North America (Blue), Asia (Light purple), Europe (Yellow) and South America (Pink).Supplementary Fig. 2
Supplementary Fig. 2 Comparison of all Indian SARS-CoV-2 genomes with Wuhan strain (first collected sample) shows variation in ORF1a and 1b protein, surface glycoprotein, ORF3a protein, ORF7a protein, ORF8 and nucleocapsid protein.Supplementary Fig. 3
Supplementary Fig. 3 Differential entropy plots for Indian vs global isolates. Mutational entropy of each amino acid position in all ORFs calculated for Indian and global isolates with respect to reference strain (Wuhan_IPBCAMS-WH-01_2019_EPI_ISL_402123). In global isolates, Indian samples have not been included to highlight differential entropy. Genomic sequences were retrieved from GISAID (https://www.gisaid.org).Acknowledgment
The seventh author (SEH) acknowledges Department of Biotechnology, Government of India for funding support (BT/PR23099/NER/95/632/2017), (BT/PR23155/NER/95/634/2017). SEH is a JC Bose National Fellow, Department of Science and Technology, Government of India & Robert Koch Fellow, Robert Koch Institute, Berlin. The first author (HS) is a recipient of Women Scientist fellowship, Department of Health Research and the second (JS) & fourth (SJ) authors received Young Scientist fellowships from the Department of Health Research, Ministry of Health and Family Welfare, Government of India. The sixth author (JAS) received UGC Startup grant and the third author (MK) received Silver Jubilee Post-Doctoral fellowship from Jamia Hamdard, New Delhi. Authors acknowledge the Originating and Submitting Laboratories for their sequences and meta-data shared through GISAID on which this study is based. Authors acknowledge BioInception Pvt. Ltd, for providing their proprietary data analysis pipeline and platform.
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- Coronavirus disease 2019 (COVID-19) Situation report-94. Geneva: WHO; 2020.
- Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility - King county, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377-81.
- [Google Scholar]
- Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: A mathematical model-based approach. Indian J Med Res. 2020;151:190-9.
- [Google Scholar]
- Severe acute respiratory illness surveillance for coronavirus disease 2019, India, 2020. Indian J Med Res. 2020;151:236-40.
- [Google Scholar]
- Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151:200-9.
- [Google Scholar]
- Molecular Analyses of Over Hundred Sixty Clinical Isolates of SARS-CoV-2: Insights on Likely Origin, Evolution and Spread, and Possible Intervention Preprints. 2020 doi: 1020944/preprints2020030320v1
- [Google Scholar]
- Emerging genetic diversity among clinical isolates of SARS-CoV-2: Lessons for today? Infect Genet Evol. 2020;84:104330. doi:10.1016/j.meegid.2020
- [Google Scholar]
- Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020;117:9241-3.
- [Google Scholar]
- Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33-46.
- [Google Scholar]
- Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33-46.
- [Google Scholar]
- Protein-protein interfaces: Analysis of amino acid conservation in homodimers. Proteins. 2001;42:108-24.
- [Google Scholar]
- An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970;132:211-50.
- [Google Scholar]





