Translate this page into:
Non-invasive prenatal rhesus D genotyping using cell-free foetal DNA
For correspondence: Dr Subhas Chandra Saha, Department of Obstetrics & Gynaecology, Postgraduate Institute of Medical Education & Research, Chandigarh 160 012, India e-mail: drscsahaa@gmail.com
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Non-invasive prenatal diagnosis (NIPD) of rhesus D (RHD) genotype using cell-free foetal DNA is extensively used in many developed countries. Studies on NIPD from India are scarce. The aim of the present study was to evaluate the performance of non-invasive foetal RHD genotyping by targeting exon 10 of the RHD gene using cell-free DNA.
Methods:
DNA was extracted from the maternal plasma of alloimmunized and non-alloimmunized women between 7 and 34 wk of gestation. RHD sequence was determined by quantitative real time polymerase chain reaction (PCR). Results were compared with RhD phenotype obtained from cord blood samples of neonates.
Results:
A total of 135 samples from RhD-negative pregnant women were collected. The foetal RHD status was conclusive in all 135 (100%) cases. The highest number of cases reported for RHD genotyping were from Punjab (38.5%) followed by Haryana (24.4%), Himachal Pradesh (17.0%) and Chandigarh Union Territory (13.3%). The non-invasive test correctly predicted the foetal RhD phenotype in 133 of 135 cases, making the accuracy of the test as 98.51 per cent [95% confidence interval (CI): 97.90-99.50%]. The overall sensitivity and specificity of the test were 99.18 per cent (95% CI: 95.52-99.98%) and 92.31 per cent (95% CI: 63.97-99.81%), respectively, with negative and positive predictive values of 99.80 per cent (95% CI: 94.85-99.87%) and 96.31 per cent (95% CI: 62.87-98.84%), respectively.
Interpretation & conclusions:
Non-invasive foetal RHD determination by single-exon quantitative PCR exhibited high accuracy and could be used in routine clinical practice after confirmatory studies are done.
Keywords
Alloimmunized
cell-free foetal DNA
exon
maternal plasma
non-invasive prenatal diagnosis
rhesus D genotyping
Since the advent of cell-free foetal DNA (cffDNA) in the maternal plasma, numerous applications have evolved in the field of molecular foetal medicine, leading a way to non-invasive prenatal diagnosis (NIPD)1. Though the relative magnitude of circulating foetal DNA is small, it has served in a copious way in determining the foetal genetic loci that are totally absent from the maternal genome. Consequently, cffDNA was explored in the field of prenatal diagnosis for the detection of foetal aneuploidy, pre-eclampsia, single gene disorders, gender determination and foetal rhesus D (RhD) genotype2. Although the use of NIPD in these applications is still in infancy stage, the feasibility of detection of foetal RhD status using circulating foetal DNA is a promising area of research.
Currently, as a major technique, non-invasive RhD genotyping by polymerase chain reaction (PCR) has revolutionized the determination of foetal RHD status in RhD-negative pregnant women, who are always on the verge of RhD alloimmunization. The alloimmunization in adverse cases destroys foetal red blood cells that lead to haemolytic disease in the foetus and newborn. However, the incidence of RhD alloimmunization in RhD-negative pregnant women was reduced considerably from 14 to 1.5 per cent by offering anti-D prophylaxis3. Before RhD genotyping, foetal RhD status was determined using an invasive procedure like chorionic villus sampling or amniocentesis, although these procedures carried a significant risk to the foetus4. The introduction of non-invasive RhD genotyping, based on the detecting of RHD gene sequences in the plasma of RhD-negative women, has obviated these risks to a larger extent.
Research groups in Western countries have carried out multiple clinical trials on NIPD. The results of these studies such as predictability, sensitivity and specificity of the test were found to be promising and reliable567. NIPD is now offered as a routine screening test for determining foetal RhD status and foetal aneuploidies in prenatal care8. However, despite the considerable advancement in non-invasive prenatal testing in other parts of the globe, the studies or clinical trials carried out in India are scanty. In the present prospective study, the clinical utility, sensitivity and specificity of the NIPD test was evaluated for foetal RhD status detection in Rh-negative pregnant women.
Material & Methods
The study was jointly conducted by the departments of Obstetrics & Gynecology and Experimental Medicine & Biotechnology, Postgraduate Institute of Medical Education & Research, Chandigarh, India, from February 2014 to August 2016. A total of 135 consecutive RhD-negative pregnant women with a mean gestational age of 23 weeks (length of gestation, 7-34 wk) were enrolled in the study. Gestational age was calculated based on menstrual cycle dates and was confirmed by ultrasound. Where gestational age was not certain, it was confirmed by ultrasound before 20 wk of gestation had passed. The participants were from different States of north India such as Punjab, Haryana, Himachal Pradesh, Chandigarh, Jammu and Kashmir, Uttar Pradesh, Uttrakhand and Bihar.
Pregnancies that had evidence of intrauterine foetal death, multiple pregnancies, foetal gross congenital malformations and the ones that had RhD-negative partner were excluded from the study. Written, informed consent was obtained from all the study participants, and the ethical clearance was obtained from the Institutional Ethics Committee that approved the study protocol (Ethics Committee approval no. 11/608).
Blood sample collection and preparation: Blood samples (10 ml) were collected from each study participant in an EDTA (ethylenediaminetetraacetic acid)-anticoagulated Vacutainer® (Becton Dickinson, Franklin Lakes, NJ, USA) and were transported the same day to the laboratory, where they were kept at +4°C and processed within 12 h. Blood samples were centrifuged at 3000 ×g for 10 min to separate out the plasma. Isolated plasma was again centrifuged at 2700 ×g for 10 min to remove the leftover residual cells. The plasma samples were aliquoted and stored at −80°C until further use.
Routine ABO and Rh typing of study participants was performed by standard agglutination methods using polyclonal as well as monoclonal antibodies in accordance with manufacturers’ recommendations (Alba Bioscience Inc., Durham, NC, and Gamma Biologicals, Houston, TX, USA).
DNA extraction and real-time polymerase chain reaction: Total DNA was extracted from 200 μl of plasma using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) as described in the manufacturer's protocol. The DNA was finally eluted with 30 μl of elution buffer. RHD gene sequence detection was performed as described elsewhere91011 with slight modification in PCR reaction conditions. The human β-globin gene was used as a positive control for total DNA extraction, and the plasma obtained from the nulligravida women carrying RhD-positive or RhD-negative blood was used as positive and negative control, respectively. Real-time PCR (RT-PCR) was performed on an ABI 7500 detection system (Applied BioSystems, USA). The RHD exon 10 consists of a forward primer, 5'-CCTCTCACTGTTGCCTGCATT-3’ and reverse primer, 5'-AGTGCCTGCGCGAACATT-3'. The β-globin primer consists of a forward primer, 5'-GT GCACCTGACTCCTGAGGAGA-3´ and reverse primer, 5'-CCTTGATACCAACCTGCCCAG-3´ (Eurofins Scientific, USA). The amplification reactions were set up in a 20 μl reaction volume, with 10 μl power SYBR® Green PCR Master Mix (Applied Biosystems) and 5 μl of template DNA and primers were added at the final concentration of 300 nM and total reaction volume were raised to 20 μl by adding nuclease-free water. DNA amplification was carried out in 96 well plates (Applied Biosystems). PCR conditions were as follows: An initial denaturation step of 10 min at 95°C, followed by amplification performed for 40 cycles of denaturation (95°C for 15 sec), annealing (58°C for 1 min) and extension (72°C for 30 sec). Melting curve analysis was carried out at the end of each PCR assay to verify the specificities of the amplified product11. Each sample was run in triplicate, and for each run, plasma obtained from nulligravida women carrying an RhD-positive or RhD-negative blood was used as positive and negative control, respectively.
Interpretation criteria: A threshold cycle (Ct) of 35-37 of positive control was considered as the threshold Ct value. A sample was considered RHD positive when a fluorescent signal was detected for the both, RHD and β-globin gene in the above-mentioned threshold range and RHD negative when a signal was detected only for the β-globin gene. The RHD gene sequence detection was considered inconclusive when the fluorescent signal obtained from the RHD reaction appeared much before the 35-37 Ct range. For that particular sample, it was assumed that the magnitude of RHD sequences detected in maternal plasma was too large to be considered of foetal origin. Samples were considered RHD-negative genotype, when all RHD PCR reactions were found negative and RHD-positive genotype when at least two out of three PCR reactions were positive. Results of prenatal RhD type were confirmed with foetal or cord blood serology or at the time of delivery.
Statistical analysis: GraphPad Prism v5.03 (GraphPad Software, CA, USA) was used for the descriptive statistical analysis. Sensitivity and specificity were calculated for RHD genotyping. All estimates were presented with 95 per cent confidence intervals (CI).
Results
At the time of blood sampling, the mean gestational age among the study participants was 23±1.3 wk. A total of 27 first trimester (wk 3-12) samples were calculated, whereas 72 and 36 samples were collected during the second (wk 13-28) and third trimesters (wk 29 and later), respectively. Of the 135 participants, 52 were from Punjab (38.51%), 33 from Haryana (24.44%), 23 from Himachal Pradesh (17.03%), 18 from Chandigarh Union Territory (13.33%), five from Jammu and Kashmir (3.70%), two from Uttar Pradesh (1.48%) and one each from Uttrakhand and Bihar accounting 1.48 per cent. About 87.40 per cent (118/135) women were RhD alloimmunized, either with significant or non-significant indirect coombs test (ICT) titres, whereas 12.59 per cent (17/135) women were those who attended the ANC with no history of alloimmunization in the present pregnancy. An ICT titres of >1:16 or <1:16 are considered as significant and non-significant, respectively, in our laboratory.
Foetal RHD exon 10 was conclusively determined in all the 135 (100%) study participants. Among these, 122 cases were genotyped as RHD positive and 13 as RHD negative. Among the 122 RHD-positive cases, one (0.81%) sample was later confirmed as false RHD positive on neonatal foetal blood serology (Figure). RHD-negative status was correctly predicted in 12 cases with one false RHD-negative case reported. Neonatal RhD status was negative in 13 (9.62%) and positive in 122 (90.37%) pregnancies. The non-invasive test correctly predicted the neonatal RhD phenotype in 133 of 135 cases, and therefore, the accuracy of the test was 98.51 per cent (95% CI: 97.9-99.5%). The overall sensitivity and specificity of the test were 99.18 per cent (95% CI: 95.52-99.98%) and 92.31 per cent (95% CI: 63.97-99.81%), respectively (Table). The negative and positive predictive values were 99.80 per cent (95% CI: 94.85-99.87%) and 96.31 per cent (95% CI: 62.87-98.84%), respectively.
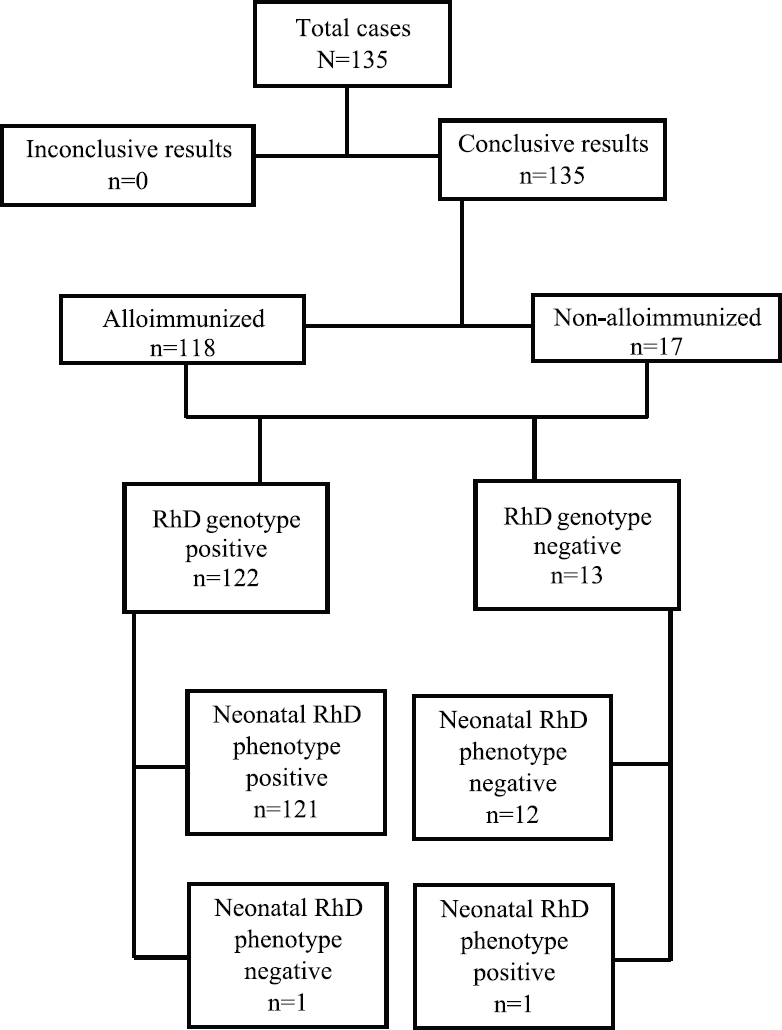
- Summary of the results of non-invasive foetal RHD genotyping in maternal plasma using cell-free foetal DNA.
| NIPD | Neonatal/foetal blood serology | Total | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|
| RhD positive | RhD negative | ||||||
| RhD positive | 121 | 1 | 122 | ||||
| RhD negative | 1 | 12 | 13 | ||||
| Total | 122 | 13 | 135 | 99.10 | 92.31 | 99.80 | 96.31 |
PPV, positive predictive value; NPV, negative predictive value; RhD, rhesus D
Discussion
Most of the studies have examined prenatal diagnostic accuracy of NIPD for foetal RHD genotyping using cffDNA of maternal plasma, ranging from 32 to 100 per cent6 taking samples from all three trimesters of pregnancy, but the majority examined second and third trimesters12. The meta-analysis by Geifman-Holtzman et al6 involving a total of 3261 samples revealed that samples taken at first trimester of pregnancy for RHD status had 90.8 per cent diagnostic accuracy, whereas 98.5 and 99 per cent accuracy were reported for samples, taken at second and third trimesters, respectively. Our study demonstrated 98.51 per cent diagnostic accuracy although 20.8, 53.3 and 26.7 per cent participants were recruited during the first, second and third trimester, respectively. A diagnostic accuracy of 96.5-100 per cent13 is considered as an acceptable reference range for this non-invasive test, for routine clinical use.
Lo et al1 in their study reported 100 and 80 per cent sensitivity and specificity, respectively whereas, another study using single exon 10 PCR on participants in their first pregnancy trimester reported both 100 per cent sensitivity and specificity9. A study, targeting exons 5 and 7 of the RHD gene reported 93 and 100 per cent sensitivity and specificity, respectively14, whereas Akolekar et al15 reported 98.2 per cent sensitivity and 100 per cent specificity in 591 women. Our data, in terms of sensitivity and specificity were consistent with above reports. Only 0.83 per cent false-positive/false-negative rate was observed in our study that might be attributed to the inclusion of the participants with higher gestational age where higher concentration of free foetal DNA exists in the maternal circulation14. A multicenter, two-exon study (exons 5 and 7), with results stratified by gestational age, reported 99.8 per cent sensitivity with 7.8 per cent inconclusive results16. Of the 4913 participants recruited, the rate of false-negative results increased significantly from 0.8 to 1.8 per cent for the participants whose sample was taken post 11 wk of gestation. Another study using exon 7 amplification strategy, in 193 first trimester Italian participants demonstrated a 93.3 per cent overall accuracy with 92.8 per cent sensitivity and 94.1 per cent specificity17. The one false-positive case in our study could be due to variant RHD type (D, DVI or pseudogene) as indicated by a low Ct value, though not confirmed genotypically whereas, another false negative was sampled at seven weeks of gestation. The less concentration of cffDNA at early gestation could be the reason for false RHD-negative results.
Although, the assay targeted a specific region (exon 10) of the RHD gene, only a single false-positive and a -negative cases were reported when compared with foetal/neonatal RhD serotype. RHD genotyping utilizing a multi-exon approach could detect appropriately some RHD variants associated with RhD-negative phenotype, thus minimizing the rate of false positives. The identification of such RHD variants depends on analysis of inconsistencies between the different targeted exons of RHD gene due to analytical variations or errors, like low cffDNA quantity during the first trimester, or faulty DNA extraction, leading to a false result1318. In clinical practice, these results would lead to the mismanagement of the RhD pregnancies who are at a risk of alloimmunization. Contrarily, targeting a single exon in replicates has shown to augment the sensitivity of foetal DNA detection in maternal plasma18.
Non-invasive prenatal screening for foetal RHD determination in sensitized as well as non-sensitized women is reported with variable sensitivity and specificity19. The clinical validity of the NIPD test remains unequivocal, regardless of the type of the individual involved. However, the magnitude of management based on NIPD may vary. For example, if the Rh blood type of the foetus is determined as RhD positive by NIPD in non-sensitized women at early gestation, such participants could be categorized as potential-risk participants. Likewise, in sensitized women, if the same test predicts the foetal blood as RhD positive, these can be directly viewed as high-risk pregnancies.
Currently, NIPD for determining the foetal RHD genotype would be suitably accurate when performed along with other tests during the antenatal visits. In non-sensitized women, this prenatal test will obviate unnecessary administration of anti-D immunoglobulin prophylaxis in women carrying the RhD-negative foetus. Making this testing coincide with other invasive or non-invasive prenatal tests may help to minimize anti-D immunoglobulin administration during pregnancy, while rapidly providing improved care to non-alloimmunized women, with reduced expenses for both, participants and the healthcare provider.
In conclusion, non-invasive RHD genotyping in early antenatal care appeared suitable for systemic screening, thus avoiding multiple visits and investigations in the event of foetal RhD-negative status. This approach also obviates the need of anti-D prophylaxis to all RhD-negative pregnant women. Further, RHD determination through single-exon analysis may prove cost-effective and safe if performed in replicates. Our data stresses the need for inclusion of NIPD testing in routine clinical diagnostic algorithm for managing Rh pregnancies, who are at a risk of alloimmunization.
Financial support & sponsorship: Authors thank the Indian Council of Medical Research, New Delhi, for providing financial support for this study.
Conflicts of Interest: None.
References
- Circulating foetal DNA: Its origin and diagnostic potential – A review. Placenta. 2004;25(Suppl A):S93-101.
- [Google Scholar]
- The scientific basis of antenatal prophylaxis. Br J Obstet Gynaecol. 1998;105(Suppl 18):11-8.
- [Google Scholar]
- Feto-maternal transfusion after chorionic villus sampling: Clinical implications. Hum Reprod. 1986;1:37-40.
- [Google Scholar]
- Antenatal prophylaxis of Rh isoimmunization: 28-weeks'-gestation service program. Can Med Assoc J. 1978;118:627-30.
- [Google Scholar]
- Diagnostic accuracy of noninvasive foetal Rh genotyping from maternal blood – A meta-analysis. Am J Obstet Gynecol. 2006;195:1163-73.
- [Google Scholar]
- Report of the first nationally implemented clinical routine screening for foetal RHD in D- pregnant women to ascertain the requirement for antenatal RhD prophylaxis. Transfusion. 2012;52:752-8.
- [Google Scholar]
- Non-invasive prenatal testing for aneuploidy and beyond: Challenges of responsible innovation in prenatal screening. Eur J Hum Genet. 2015;23:1592.
- [Google Scholar]
- Foetal RHD genotyping in maternal serum during the first trimester of pregnancy. Br J Haematol. 2002;119:255-60.
- [Google Scholar]
- Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033-5.
- [Google Scholar]
- The most favourable procedure for the isolation of cell-free DNA from the plasma of iso-immunized RHD-negative pregnant women. J Circ Biomark. 2015;4:12.
- [Google Scholar]
- Noninvasive prenatal diagnosis of foetal blood group phenotypes: Current practice and future prospects. Prenat Diagn. 2009;29:101-7.
- [Google Scholar]
- Diagnostic accuracy of non-invasive foetal RhD genotyping using cell-free foetal DNA: A meta analysis. J Matern Foetal Neonatal Med. 2014;27:1839-44.
- [Google Scholar]
- Non-invasive foetal RHD genotyping in the first trimester of pregnancy. Clin Chem Lab Med. 2010;48:1121-6.
- [Google Scholar]
- Foetal RHD genotyping in maternal plasma at 11-13 weeks of gestation. Foetal Diagn Ther. 2011;29:301-6.
- [Google Scholar]
- Accurate and robust quantification of circulating foetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clin Chem. 2005;51:312-20.
- [Google Scholar]
- Diagnostic accuracy of routine antenatal determination of foetal RHD status across gestation: Population based cohort study. BMJ. 2014;349:g5243.
- [Google Scholar]
- Non-invasive prenatal RHD genotyping using cell-free foetal DNA from maternal plasma: An Italian experience. Transfus Med Hemother. 2015;42:22-8.
- [Google Scholar]
- Replicate real-time PCR testing of DNA in maternal plasma increases the sensitivity of non-invasive foetal sex determination. Prenat Diagn. 2003;23:235-8.
- [Google Scholar]






