Translate this page into:
Mismatch amplification mutation assay-polymerase chain reaction: A method of detecting fluoroquinolone resistance mechanism in bacterial pathogens
For correspondence: Dr Indrani Karunasagar, Nitte University Centre for Science Education & Research, Kotekar-Beeri Road, Paneer Campus, Derelakatte, Mangaluru 575 018, Karnataka, India e-mail: indrani.karunasagar@nitte.edu.in
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The mismatch amplification assay is a modified version of polymerase chain reaction (PCR) that permits specific amplification of gene sequences with single base pair change. The basis of the technique relies on primer designing. The single nucleotide mismatch at the 3’ proximity of the reverse oligonucleotide primer makes Taq DNA polymerase unable to carry out extension process. Thus, the primers produce a PCR fragment in the wild type, whereas it is not possible to yield a product with a mutation at the site covered by the mismatch positions on the mismatch amplification mutation assay (MAMA) primer from any gene. The technique offers several advantages over other molecular methods, such as PCR-restriction fragment length polymorphism (RFLP) and oligonucleotide hybridization, which is routinely used in the detection of known point mutations. Since multiple point mutations in the quinolone resistance determining region play a major role in high-level fluoroquinolone resistance in Gram-negative bacteria, the MAMA-PCR technique is preferred for detecting these mutations over PCR-RFLP and sequencing technology.
Keywords
Fluoroquinolone resistance
mismatch amplification mutation assay
polymerase chain reaction
quinolone resistance determining region mutations
Introduction
The PCR is a powerful standard tool for the in vitro amplification of specific DNA sequences in molecular biology. In the allele-specific PCR (AS-PCR) the modification allows the specific amplification of DNA sequences with single-point mutation with the help of reverse primer harbouring a mismatch at the 3’ end; hence, it is also widely known as mismatch amplification mutation assay-PCR (MAMA-PCR). The technique was well established and experimented in the late 1980s in the detection of point mutation of several disease conditions, such as German familial amyloidotic polyneuropathy type II1 and sickle cell anemia2. Over the years, many methods have been developed to detect such mutations34. The MAMA-PCR technique is also widely used for the detection of point mutations in the quinolone resistance determining regions (QRDRs) of fluoroquinolone-resistant bacterial pathogens5678.
Fluoroquinolones are the drug of choice for most of the urinary tract infections caused by bacterial pathogens. However, resistance to these drugs has been frequently reported and further resulted in treatment failures. Fluoroquinolones are synthetic class of antibacterials that act on both DNA gyrase and topoisomerase IV and have many clinical applications. Topoisomerases play a prominent role in DNA replication where DNA gyrase or topoisomerase II (2 gyrA and 2 gyrB subunits) introduces negative supercoils into DNA and topoisomerase IV (2 parC and 2 parE subunits) responsible for removing the interlinking structure between the two newly produced DNA strands during replication9. Mutations in the QRDR of bacterial pathogens inhibit the binding of fluoroquinolone to DNA and enzyme (topoisomerase targets) complex and are the major cause of fluoroquinolone resistance. Hence, it is important to identify these mutations to introduce suitable therapeutic strategies in severe bacterial infections. Several point mutations in QRDR have been attributed to fluoroquinolone resistance in bacterial pathogens. These mutations are found most frequently in the Ser 83 and Asp 87 codons of the gyrA and Ser 80 and Glu 84 codons of parC in Escherichia coli as well as in other microorganisms101112. Although sequencing technique is the most appropriate method for identifying point mutations in any gene, its high cost and extensive process have led to the development of cost-effective methods such as MAMA-PCR and PCR-restriction fragment length polymorphism (RFLP). The modified MAMA techniques have also been successfully applied to viruses13 and eukaryotic species14. However, the specificity of oligonucleotide primers designed for MAMA-PCR is still a challenge for the development of this protocol for use in disease diagnosis.
Principle of MAMA-PCR
MAMA-PCR works on the principle of standard PCR in which amplification of target DNA sequences is mainly facilitated by the oligonucleotide reverse primer with single nucleotide mismatch at the 3’ end. A single primer-template mismatch at the 3’ proximity of oligonucleotide primer just before the last nucleotide will have little or no effect on the amplification of genes during PCR provided there is no mutation in the target gene. Thus, wild-type genes can be amplified efficiently6. However, a point mutation in the target gene will create additional mismatch with the oligonucleotide primer during the amplification process. The mismatch generated at the 3’-OH end makes Taq DNA polymerase unable to extend the primer for further amplification. Hence, the gene with point a mutation generates double mismatch at 3’-OH end is responsible for inhibition of the PCR amplification process15 (Fig. 1).
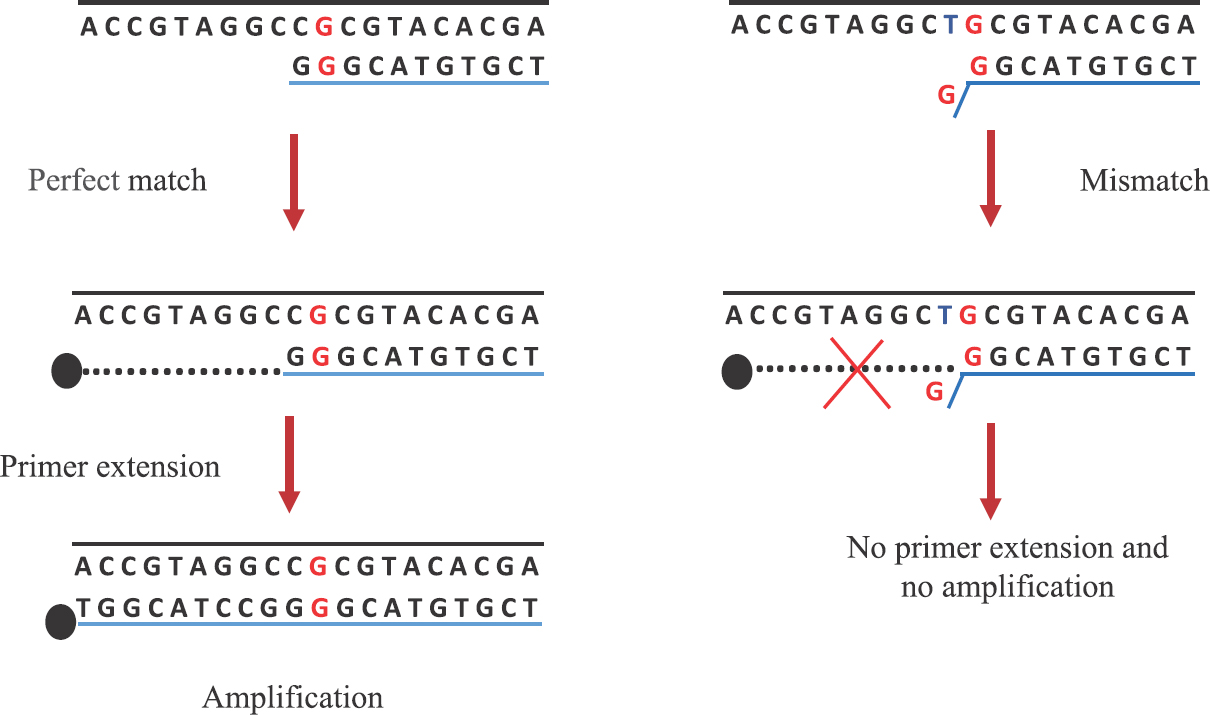
- Principle of the mismatch amplification mutation assay-polymerase chain reaction.
Primer designing protocol
The MAMA-PCR primers can be designed either to amplify the region with specific mutations and no amplification for wild types516 or wild types can be amplified and no PCR product can be generated in the case of point mutations at specific sites617. The primers are carefully designed considering all aspects such as specificity and sensitivity of the technique using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). One forward primer and two reverse primers are used for PCR amplification. The normal forward and reverse primers are used to amplify the gene in both mutant and wild types and the second reverse primer will act as a MAMA reverse primer with single nucleotide mismatch at 3’ end (Fig. 2). For detecting QRDR mutations, the method usually follows a duplex PCR technique wherein two bands can be obtained in wild type and a single band can be seen in mutant version same gene9.
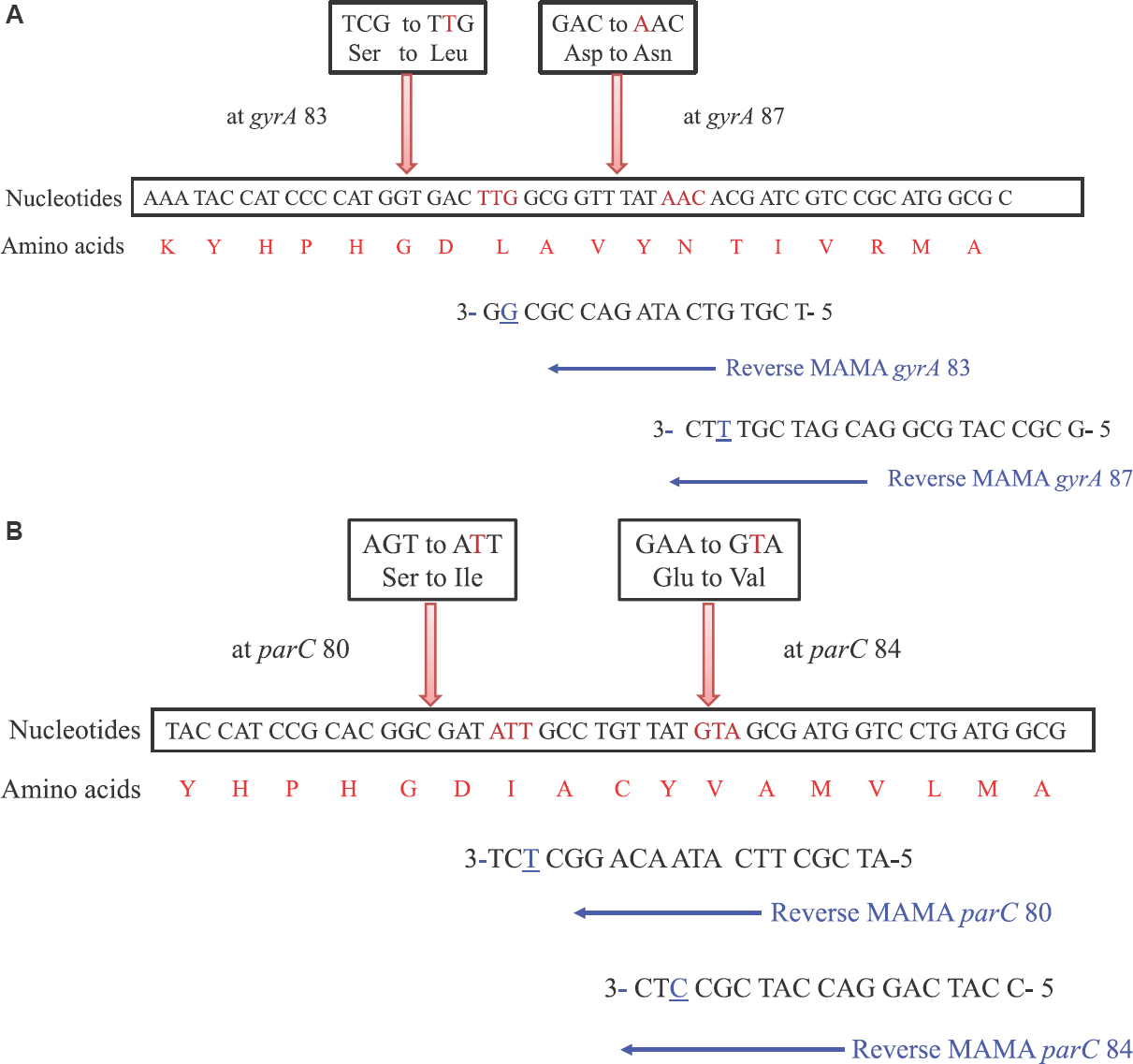
- Illustration of mismatch amplification mutation assay-polymerase chain reaction primers for gyrA (A) and parC (B) mutation detection. The underlined nucleotides are mismatched nucleotides at 3’ end of each mismatch amplification mutation assay primer. The amino acid found in the native protein and the changed amino acid due to point mutation are indicated above the corresponding nucleotide sequences.
Preparation of bacterial DNA and MAMA-PCR protocol
Bacterial DNA isolation can be done by using cetyl trimethyl ammonium bromide (CTAB) method described by Ausubel18. The quantification of DNA is generally performed by spectrophotometric measurement of the absorption at 260 nm.
A common MAMA-PCR 30 μl volumes of master mix contains 3 μl of 10× Taq buffer, 83 μM of each of the four deoxyribonucleotide triphosphates, 30 pmol of forward primer, 20 pmol of MAMA reverse primer, 10 pmol control reverse primer and 1 U of Taq DNA polymerase with 2 μl of template DNA. The MAMA-PCR cycling conditions commonly starts with an initial dissociation step at 95°C for 5 min to ensure the complete separation of the DNA strands followed by initial denaturation at 95°C for 5 min and 35 cycles of denaturation at 94°C, annealing at required temperature (55-65°C) and extension at 72°C each for 40 sec, respectively. A final extension step of 5-10 min at 72°C ensures complete extension of all amplicons. To measure the success of PCR amplification, 8-10 μl of PCR product with gel loading buffer is run on 2 per cent agarose gel pre-stained with ethidium bromide (0.5 μg/ml) in 1× TAE buffer and observed under ultraviolet light in a gel documentation system to confirm the results.
Modifications of MAMA-PCR
Melt MAMA-PCR is the modification of MAMA-PCR that utilizes labelled two AS primers with a mismatch at the 3’ end. One of the AS primers also differs with GC clamp at the 5’ end to increase the melting temperature of the corresponding amplicons to enable the easy differentiation of AS-PCR products through melt curve analysis19.
AS probe and primer amplification assay-PCR technique uses four AS TaqMan MGB probes along with mismatched AS primers and a common forward primer. These primers selectively amplify each allele in independent runs of real-time PCR, resulting in fluorescence corresponding to the increased DNA concentration20.
AS blocker-PCR is an improvement of AS-PCR so that amplification of primer-template mismatches would be suppressed. The shortened mutant AS primer at the 5’ end reduced the Tm (melting temperature) and a second blocking oligonucleotide is complementary to the wild type but phosphorylated at 3’ end to prevent extension and suppresses non-specific amplification of wild types with AS primers21.
Comparison of MAMA-PCR with other methods
MAMA-PCR is one of the rapid, simple and cost-effective methods for the detection of known single-point mutations in the QRDRs of fluoroquinolone resistance bacterial pathogens. Although the methods such as gene sequencing and PCR-RFLP can be used for the detection and analysis of point mutations, the importance of MAMA-PCR over gene sequencing can be justified when a large number of bacterial isolates need to be screened for resistance characteristics during epidemiologic investigations. Further, PCR-RFLP can be used only if the point mutations generate or abolish the restriction site for the available restriction enzymes. A precise comparison of all these methods is shown in Table.
| Methods | Concept | Advantages | Limitations |
|---|---|---|---|
| MAMA-PCR | An oligonucleotide with single mismatch binds weakly to its imperfect complement | Single step, rapid, sensitive and specific method for single base pair substitution, can used to screen large number of samples | Can be used to identify only the known mutations, a specific primer is needed for detecting each mutation |
| PCR-RFLP | Two-step method involving uniplex PCR followed by restriction fragment length polymorphism | Two-step, sensitive and specific method, utilizes restriction enzymes | Depends on the efficiency of restriction enzyme digestion, all possible point mutations cannot be detected due to the absence of restriction sites |
| DNA sequencing | Process of determining the precise order of nucleotides using automated techniques | More accurate, specific, enables the detection of mutation at unknown site | Cost and time intensive |
MAMA-PCR, mismatch amplification mutation assay-polymerase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism
Conclusion
Emergence of antimicrobial resistance among bacterial pathogens is a cause of concern and can be a serious public health threat. Point mutations in the specific genes are found to be the main reason responsible for the increased resistance to some otherwise potent antimicrobial agents in microorganisms. Hence, the MAMA-PCR technique may serve as a powerful tool to identify these point mutations in a large population of microorganisms.
Financial support & sponsorship: The last author (IK) acknowledges Nitte (Deemed to be University) intramural research fund and Indian Council of Medical Research, New Delhi, for financial support (AMR/37/2011-ECD-1).
Conflicts of Interest: None.
References
- Direct sequencing of the gene for Maryland/German familial amyloidotic polyneuropathy type II and genotyping by allele-specific enzymatic amplification. Genomics. 1989;5:535-40.
- [Google Scholar]
- Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci U S A. 1989;86:2757-60.
- [Google Scholar]
- Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667-70.
- [Google Scholar]
- 4-quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:387-9.
- [Google Scholar]
- Ciprofloxacin resistance in Campylobacter jejuni isolates: Detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999;37:3276-80.
- [Google Scholar]
- Molecular analysis of ciprofloxacin resistance mechanisms in Malaysian ESBL-producing Klebsiella pneumoniae isolates and development of mismatch amplification mutation assays (MAMA) for rapid detection of gyrA and parC mutations. Biomed Res Int. 2014;2014:601630.
- [Google Scholar]
- Genotyping and antibiotic resistance of thermophilic campylobacter isolated from chicken and pig meat in Vietnam. Gut Pathog. 2016;8:19.
- [Google Scholar]
- Development of multiplex-mismatch amplification mutation-PCR assay for simultaneous detection of Campylobacter jejuni and mutation in gyrA gene related to fluoroquinolone resistance. Foodborne Pathog Dis. 2016;13:642-5.
- [Google Scholar]
- Comparison of mismatch amplification mutation assay PCR and PCR-restriction fragment length polymorphism for detection of major mutations in gyrA and parC of Escherichia coli associated with fluoroquinolone resistance. Microb Drug Resist. 2019;25:23-31.
- [Google Scholar]
- Characterization of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli by DNA sequence analysis and MAMA PCR. FEMS Microbiol Lett. 2000;190:1-7.
- [Google Scholar]
- Differential expression of virulence genes and role of gyrA mutations in quinolone resistant and susceptible strains of Salmonella weltevreden and Newport isolated from seafood. J Appl Microbiol. 2015;119:970-80.
- [Google Scholar]
- Multiple antimicrobial resistance and novel point mutation in fluoroquinolone-resistant Escherichia coli isolates from Mangalore, India. Microb Drug Resist. 2017;23:994-1001.
- [Google Scholar]
- Specific detection of naturally occurring hepatitis C virus mutants with resistance to telaprevir and boceprevir (protease inhibitors) among treatment-naïve infected individuals. J Clin Microbiol. 2012;50:281-7.
- [Google Scholar]
- Individual single tube genotyping and DNA pooling by allele-specific PCR to uncover associations of polymorphisms with complex diseases. Clin Chim Acta. 2007;376:155-62.
- [Google Scholar]
- Effects of primer-template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999-1005.
- [Google Scholar]
- Comparison of mismatch amplification mutation assay with DNA sequencing for characterization of fluoroquinolone resistance in Neisseria gonorrhoeae. J Clin Microbiol. 2004;42:591-4.
- [Google Scholar]
- Use of a rapid mismatch PCR method to detect gyrA and parC mutations in ciprofloxacin-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 2002;49:549-52.
- [Google Scholar]
- Current protocols in molecular biology. New York: Current Protocols; 1994. p. :2-4.
- [Google Scholar]
- Interleukin-28B genotyping by melt-mismatch amplification mutation assay PCR analysis using single nucleotide polymorphisms rs12979860 and rs8099917, a useful tool for prediction of therapy response in hepatitis C patients. J Clin Microbiol. 2011;49:2706-10.
- [Google Scholar]
- The allele-specific probe and primer amplification assay, a new real-time PCR method for fine quantification of single-nucleotide polymorphisms in pooled DNA. Appl Environ Microbiol. 2012;78:1063-8.
- [Google Scholar]
- Mutation detection by real-time PCR: A simple, robust and highly selective method. PLoS One. 2009;4:e4584.
- [Google Scholar]






