Translate this page into:
Candidate gene polymorphisms related to lipid metabolism in Asian Indians living in Durban, South Africa
For correspondence: Dr Datshana P. Naidoo, Department of Cardiology (Medical School), University of KwaZulu Natal, Private Bag 7, Congella, Durban 4013, South Africa e-mail: naidood@ukzn.ac.za
-
Received: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Asian Indians have been shown to have a high prevalence of metabolic syndrome (MetS), related to insulin resistance and possibly genetic factors. The aim of this study was to determine the genetic patterns associated with MetS in Asian Indians living in Durban, South Africa.
Methods:
Nine hundred and ninety nine participants from the Phoenix Lifestyle Project underwent clinical, biochemical and genetic assessment. MetS was diagnosed according to the harmonized definition. The apolipoprotein A5 Q139X, lipoprotein lipase (LPL) Hinf I, human paraoxonase 1 (PON1) 192Arg/Gln, cholesteryl ester transfer protein (CETP) Taq1B, adiponectin 45T>G and leptin (LEP) 25CAG were genotyped by real-time polymerase chain reaction in participants with and without MetS. Univariate-unadjusted and multivariate-adjusted relations were conducted for all analyses.
Results:
The prevalence of MetS was high (49.0%). More females had MetS than males (51.0 vs 42.8%). There was no significant difference in the distribution of genotypes between participants with MetS and those without. Males with the MetS who had the adiponectin TG genotype and human paraoxonase 1 AA genotype were more likely to have reduced high-density lipoprotein cholesterol (HDL-C) (P=0.001) and higher systolic blood pressure (P=0.018), respectively.
Interpretation & conclusions:
About half of the Asian Indians living in Phoenix had MetS. No association between the polymorphisms studied and the risk for MetS was observed. The adiponectin TG genotype may be associated with reduced HDL-C and the human paraoxonase 1 AA genotype with hypertension in males. This suggested that lifestyle factors were the major determinant for MetS in this ethnic group and the genetic risk might be related to its component risk factors than to MetS as an entity.
Keywords
Dyslipidaemia
genotype
hypertension
insulin resistance
metabolic syndrome
single-nucleotide polymorphism
Insulin resistance (IR) and obesity contribute to the development of risk factor clustering in the metabolic syndrome (MetS). Several investigators have examined the prevalence of MetS across different ethnic groups. Certain ethnic groups such as Asian Indians have been found to be more predisposed to developing the MetS12. In addition to IR and obesity, early evidence from twin and familial aggregation studies3 has suggested a genetic contribution to the pathogenesis of the MetS. Genome-wide scans have identified various chromosomal regions with suggestive linkage to the MetS4. To date, varying associations have been reported between single-nucleotide polymorphism(s) (SNPs) of certain genes and the MetS5. A genetic predisposition might explain the increased susceptibility to cardiovascular disease in South African Indians6. This study was undertaken to evaluate SNPs in selected candidate genes associated with lipid and carbohydrate metabolism in Asian Indians living in Durban, South Africa.
Material & Methods
All participants from the Phoenix Lifestyle Project (PLP) (Ethical reference: BE336/05) who consented to genetic screening were included in this study. The study design, the randomization process and the risk factor profile of this sample have been previously published7. Briefly, the PLP was a cross-sectional study (conducted from January 2007 to December 2008) of 1428 South African Indians (aged 15 to 64 yr) living in the cadastral area of Phoenix, Durban, South Africa. Participants for the PLP were selected randomly from the previous population census, and using the Kish method, one participant from each home was selected7.
Where written consent to participate was obtained, and before the test days, specially trained fieldworkers interviewed participants and all demographic data (sex, age, education and income level, including physical activity, diet, smoking habits, alcohol consumption, history of diabetes mellitus, hypertension) and cardiovascular risk factors were recorded in the STEPS instrument for non-communicable disease (NCD) risk factors, a modified version 1.48. The genetic study protocol was reviewed by the Biomedical Research Ethics Committee, University of Kwa-Zulu Natal, South Africa (Ethical reference: BE232/010).
Study population: A total of 999 South African Indians (mean age: 45.4±13.1 yr), comprising 749 females (mean age: 46.0±12.3 yr) and 250 males (43.4±15.2 yr), who consented to genetic screening were enrolled in this study. Anthropometric, physiological and biochemical parameters were recorded in the STEPS instrument for NCD risk factors (version 1.4)8. The clinical evaluation was conducted at the Lifestyle Centre, Inkosi Albert Luthuli Central Hospital, Durban, South Africa. Anthropometric measurements included waist circumference, weight and height (as per the WHO criteria)8. Blood pressure (BP) readings were recorded at two-minute intervals (average of three readings was recorded). Systemic hypertension was diagnosed if individuals reported hypertension and/or had readings >140 and >90 mmHg and/or on antihypertensive therapy. After an overnight fast, venous blood (20 ml) was drawn for measuring serum lipid [total triglyceride, high-density lipoprotein cholesterol (HDL-C) and total cholesterol], serum insulin and plasma glucose levels. Plasma insulin was measured by immunoassay, and glucose oxidase method was used to measure fasting plasma glucose7. Blood samples (20 ml) for the genetic analysis (Roche, South Africa) were collected in ethylenediaminetetraacetic acid tubes (EDTA).
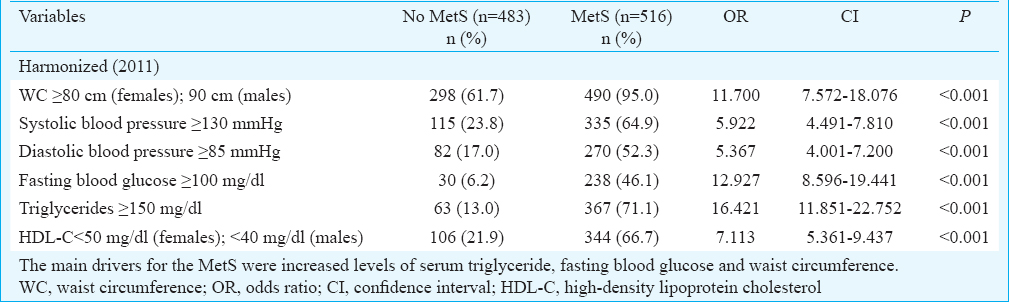
The diagnosis of the MetS was in accordance with the harmonized definition using the ethnic-specific cut-offs for waist circumference in Asian participants9. Participants with the MetS have ≥3 of 5 metabolic risk factors, viz. (i) central obesity: waist circumference ≥90 cm in males or ≥80 cm in females; (ii) triglycerides: ≥150 mg/dl; (iii) HDL-C: <40 mg/dl in males or <50 mg/dl in females; (iv) BP: ≥130/85 mmHg; and (v) fasting blood glucose: ≥100 mg/dl.
DNA extraction: Genomic DNA was extracted from whole blood using the MagNA Pure Instrument and a MagNA Pure LC Total Nucleic Acid Isolation Kit according to the manufacturer's instructions (Roche, South Africa). Briefly, 200 μl of whole blood was transferred to the sample cartridge and loaded onto the MagNA Pure LC workstation together with the necessary disposables and kit reagents. The MagNA Pure LC (Roche, South Africa) used magnetic bead technology and automatically performed the isolation and purification steps, binding of DNA, washing steps and elution of the nucleic acid. DNA concentrations were then determined using the NanoDrop 1000 analyzer (Thermo Scientific, USA) and samples were standardized to 5 ng/μl.
Selection and genotyping of polymorphisms: Six SNPs related to lipid metabolism (apolipoprotein Q139X - rs121917821), IR [cholesterol ester transfer protein Taq1B - rs708272; lipoprotein lipase (LPL) Hinf I - rs328; paraoxonase 1 192Arg/Gln - rs662] and obesity (leptin 25CAG - rs104894023; adiponectin 45T>G - rs2241766) were selected. Gene SNPs chosen were relevant to lipid metabolism and the risk for the MetS. The SNP database (dnSNP) at NCBI (http://www.ncbi.nlm.nih.gov/snp) was used. The selection of selected SNPs was based on the following: (i) In a study of 200 participants with an allelic frequency of <0.25 per cent, the apolipoprotein A5 (APOA5) Q139X showed significant associations with dyslipidaemia10. Asian Indians in South Africa have been shown to have a high prevalence of dyslipidaemia6. We predicted that the APOA5 Q139X SNP may be related to an increased risk of dyslipidaemia and thereby the MetS. (ii) The LPL (lipoprotein lipase) HinfI SNP has been linked with increased lipolytic activity and dyslipidaemia11. Since the allelic frequency for the LPL HinfI varied amongst different ethnicities1213 and this SNP had been examined in South African Indians with myocardial infarction (MI)6, we hypothesized that this SNP might be related to an increased risk of the MetS in Asian Indians. (iii) The high prevalence of an atherogenic lipid profile and diabetes amongst Asian Indians14 suggested a possible genetic risk for the MetS. We selected the PON1 (paraxonase 1) SNPs since the 192Arg allele has been shown to be associated with paraoxonase 1 activity and varying affinity for HDL15. (iv) Since it has been shown that Asian Indians with MI have low HDL-C levels6, we selected the CETP (cholesteryl ester transfer protein) Taq1B SNP for study which has revealed associations with high CETP activity and reduced HDL-C levels16. (v) In Asian individuals, the ADP (adiponectin) +45T allele has been found to be associated with IR17, which is known to be the driving factor for the MetS. We, therefore, investigated whether the +45T>G SNP was associated with the MetS in our sample. (vi) In keeping with other studies on Asian Indians18, our previous study showed that obesity was a driving factor for the MetS in our sample7. We selected the leptin (LEP) 25CAG SNP since it has been shown to be associated with obesity19.
Genotyping
Genotyping of the selected SNPs was carried out using polymerase chain reaction (PCR) (probe-specific) on the LightCycler 480 (Roche, South Africa). Amplification of the genomic DNA was obtained using a 13 μl volume, which contained PCR grade water (Roche), genotyping master mix (Roche), forward and reverse primers, forward and reverse probes (Roche), MgCl2 (where necessary) and 5 μl genomic DNA template. Primer and probe [designed by Roche, South Africa, using the LC Probe Design Software 2.0. using a reference sequence for each of the selected genes from OMIM (https://www.omim.org)] sequences are shown in Table I.
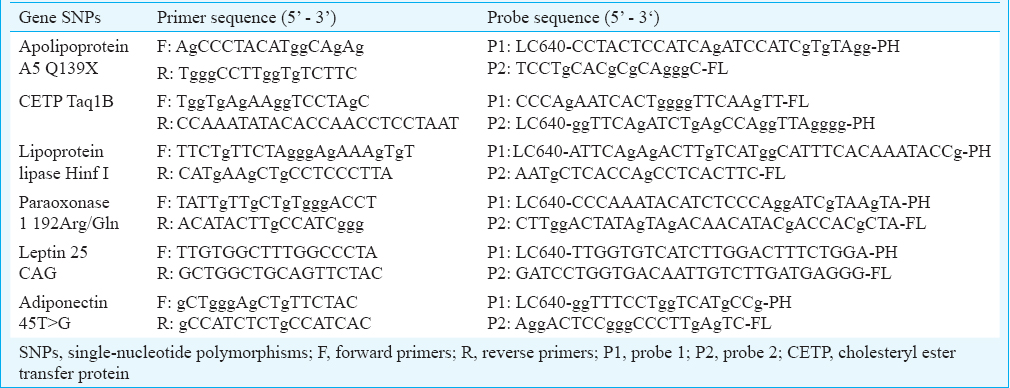
PCR occurred by denaturation at 95°C for 10 sec, annealing at 55°C for 10 sec and extension/elongation at 72°C for 10 sec. On completion of the amplification, a melting curve step occurred by cooling and reheating the PCR mix at 45°C and 95°C, respectively. Final cooling occurred for each PCR mix at 40°C for 30 sec. The integrity of the PCR products was checked in five per cent of randomly selected samples by standard techniques using the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare, Illinois, USA). Purified PCR products were run on a 1.5 per cent agarose gel and visualized using GelVue UV Transilluminator (SynGene, London, UK). To confirm the PCR findings, sequencing was performed on five per cent of the samples using the Sanger method by standard techniques.
Power calculation: A minimum f of 0.1 was detected with a sample size of 999 participants with 5 per cent significance and 90 per cent power. When HWE holds, χ2 has a Chi-square distribution with 1 df. When HWE does not hold, χ2 has a non-central Chi-square distribution with non-centrality parameter nf2. The cut-off for significance at the 5 per cent level of Chi-square with 1 df was 3.84. Thus, P value was <0.05 if a test statistic greater than 3.84 was observed. To be at least 90 per cent sure of rejecting HWE, when HWE was false, the non-centrality parameter should be at least 10.51.
Statistical analysis: Data were analyzed using the Stata 13.0 (StataCorp LP, USA). The frequencies for the studied SNPs were calculated by gene counting, and the Pearson Chi-square (χ2) test was used to identify departure from Hardy-Weinberg equilibrium (HWE). The Pearson χ2 test was also used to test associations between independent categorical variables and MetS. If an expected cell count contained fewer than five observations, then the Fisher's exact test was employed. Difference by means of MetS components and SNPs was assessed using one-way analysis of variance (ANOVA). If the data were not normal, then the Kruskal-Wallis test was used. Adjustment for multiple testing was performed using the Bonferroni correction (calculated using “qqvalue” ado in Stata 13.0). Bivariate and multivariable ordered logistic regression analyses were performed to assess the association of various MetS parameters with the observed genotypes among MetS versus non-MetS individuals.
Results
There was a high prevalence of MetS (49.0%) with a slightly higher prevalence in females (n=382/749, 51.0%) than males (n=107/250, 42.8%). The most frequent risk factor components in participants with the MetS were increased waist circumference (95.0%) and elevated triglycerides (71.1%). Increased waist circumference was also present in 61.7 per cent of individuals without MetS (Table II). The prevalence of increased triglyceride was 5.5-fold and fasting blood glucose 7.4-fold in those with MetS.
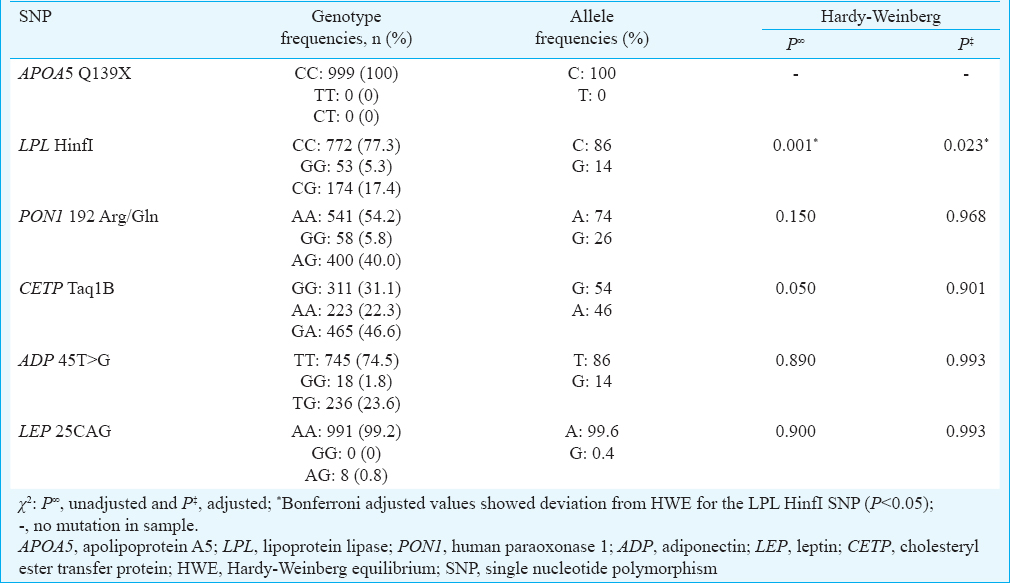
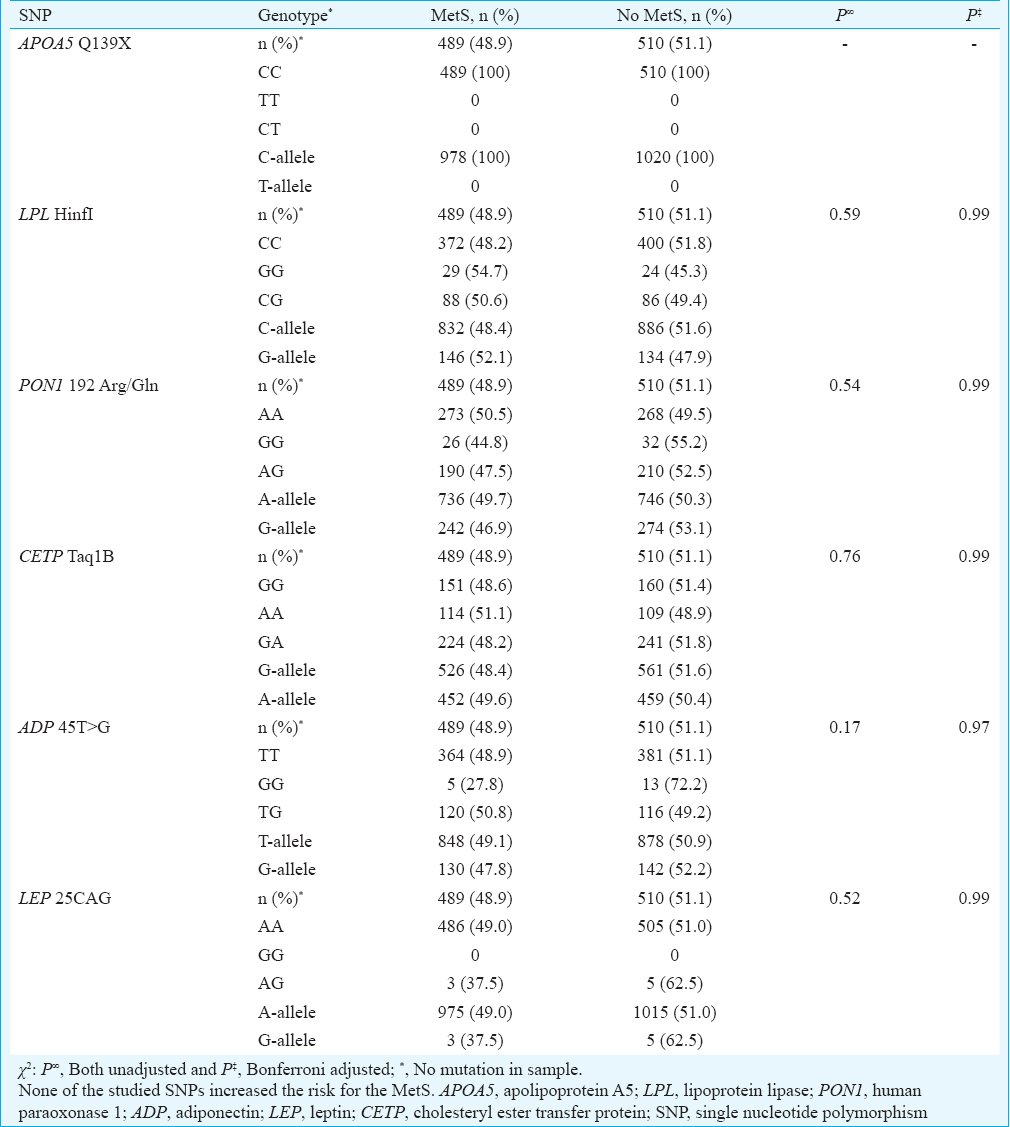
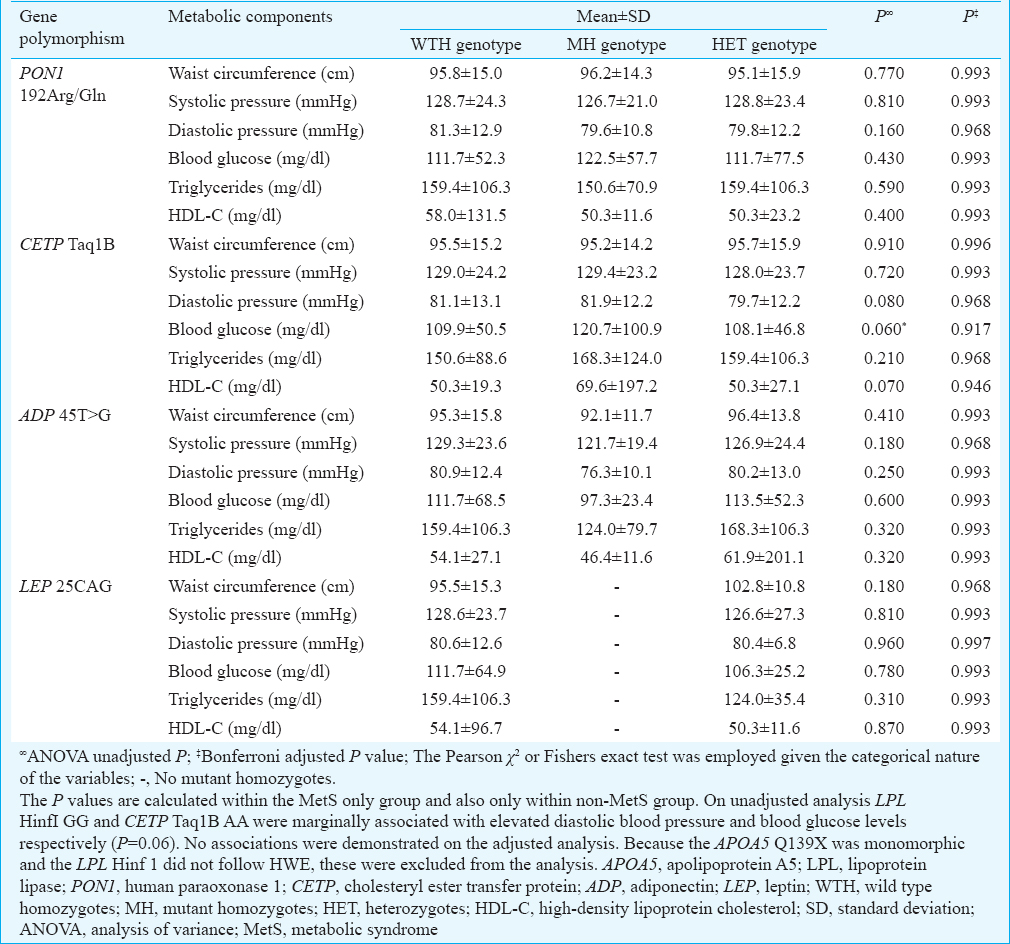
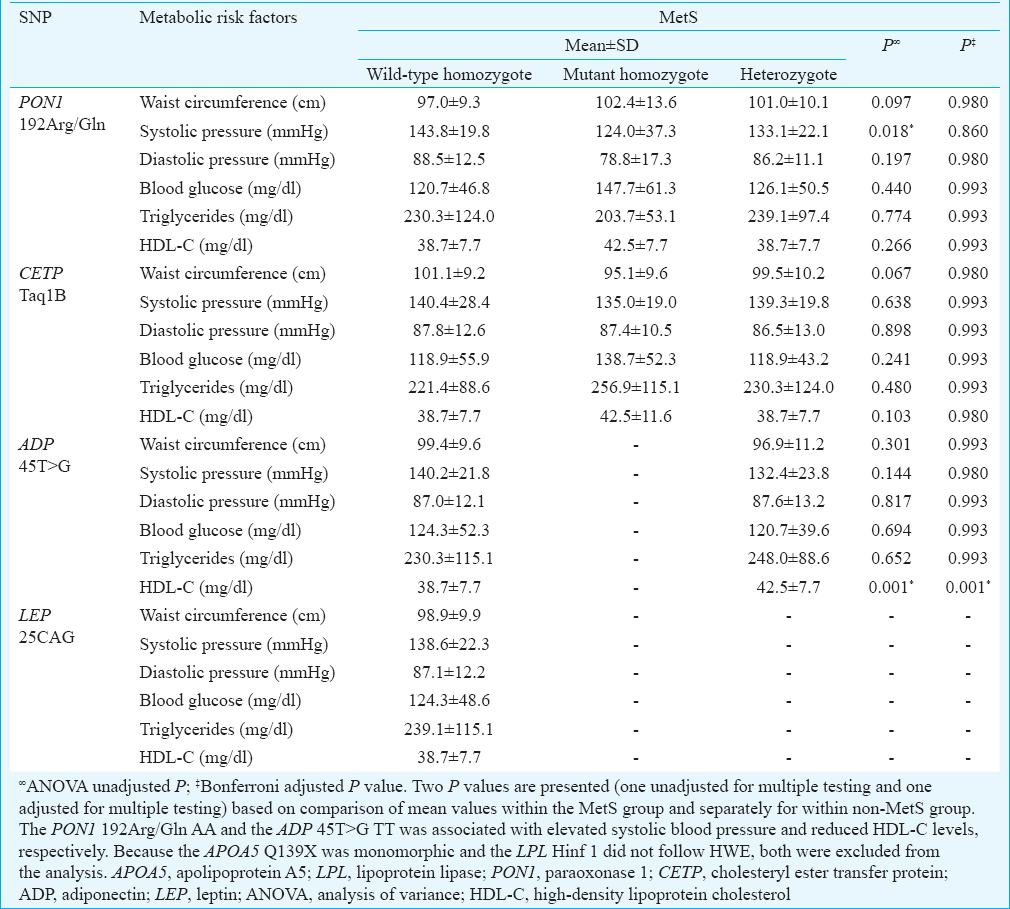
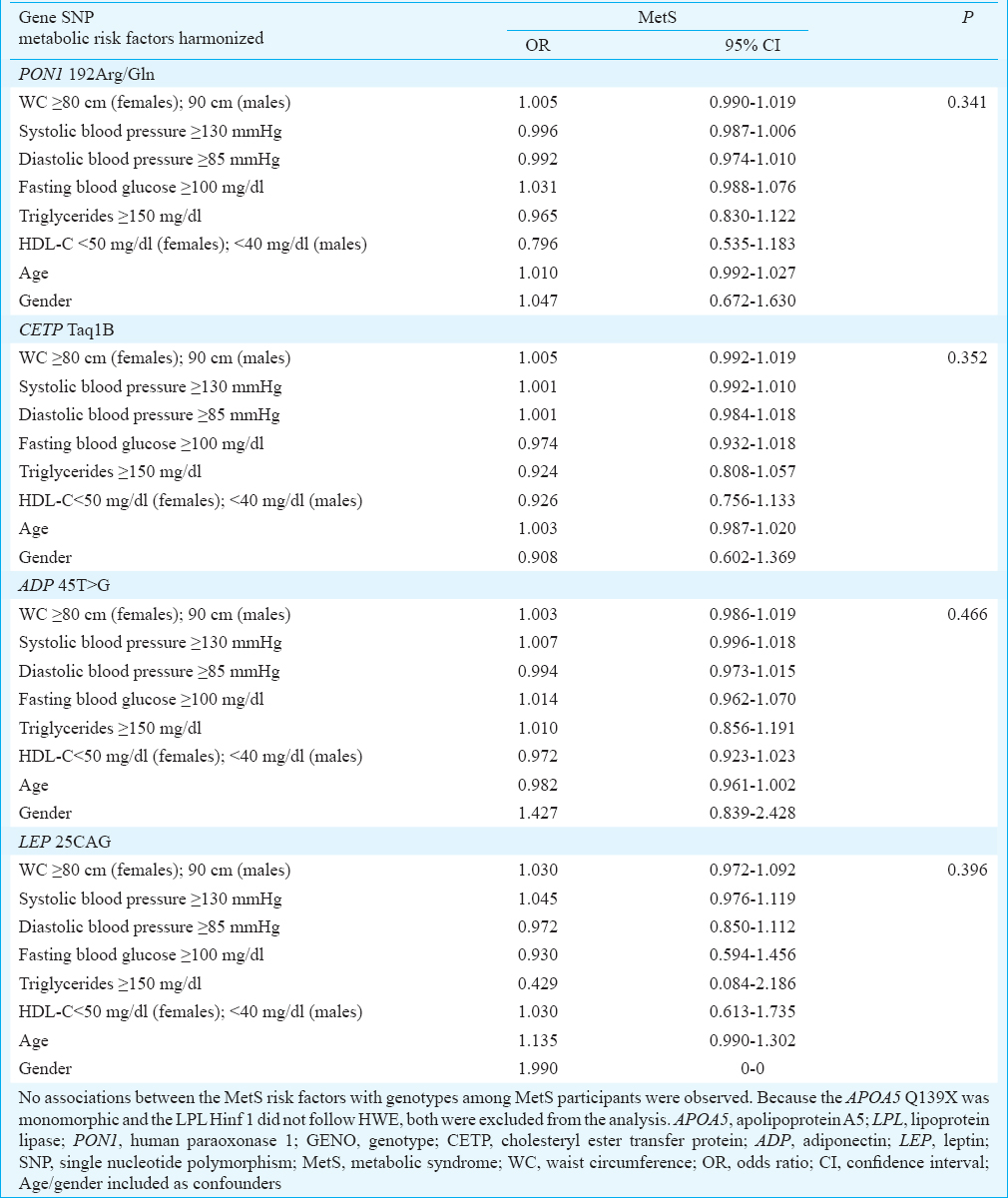
The genotype distributions of the six SNPs were in HWE, except the LPL Hinf I which deviated from HWE (Table III). Comparison of genotype and allele distribution revealed that none of the studied SNPs were significantly associated with the MetS (Table IV). The risk factor components of the MetS for all participants were compared with the genotypes of the six SNPs (Table V). In addition, the MetS risk factor components were compared with the genotype/allele of the six SNPs in males and in females to look for gender associations (Table VI). The 45T>G SNP of adiponectin was associated with reduced HDL-C levels in male participants with the MetS (P=0.001). These findings remain significant on adjusted analysis. The 192Arg/Gln SNP of human paraoxonase 1 was associated with elevated systolic BP in male participants with the MetS (P=0.018) (Table VI), but this significance fell away on adjusted analysis. No genotype/gender associations were observed in females. Similarly, no associations between the MetS risk factors with genotypes amongst MetS participants were observed (Table VII).
Discussion
In this study of Asian Indians, no genetic predisposition to the MetS was observed for the selected SNPs. While studies performed in South Asians have shown positive associations between the FABP2Ala54Thr20, APOCIII T-455C and APOCIII C-482T21, APOA1 T655C, APOA1 T756C and APOA1 T1001C SNPs22 and the MetS, only a few studies have been performed in Asian Indians. Studies that have examined that the IRS-1 G-972R, PPAR-Gamma P12A, PPAR-Gamma KCNJIIE23K, TNF-Alpha-308G/A23, PPAR-Gamma C1A and PPAR-Gamma UCP124 have failed to show any association with the MetS.
Associations have been described between the selected candidate gene SNPs and IR. For example, carriers of the CETP Taq1B in Spanish25 and the adiponectin 45T>G (+45T allele) in Taiwanese have been associated with IR26. This is probable since IR is the driving factor for the development of the MetS.
Our study was adequately powered to detect differences in genotype frequencies. It should be noted that studies showing positive associations between the SNPs we studied and the MetS have been performed on <100 study samples compared to ours. For example, the APOA5 Q139X SNP was evaluated in nine White participants10, the LPL HinfI in 99 Caucasians27 and the leptin 25CAG in 30 Thai participants19. It is also possible that population stratification could have been a confounding factor, leading to false-positive findings in studies with small samples.
A low prevalence of the MetS has been described in Singapore Indians (20.9% of males and 15.5% of females)28 and in Indians from Mauritius (10.6% in males and 14.7% in females)29. Our findings were in keeping with high prevalence described in European Asian Indians (46.0% of males and 38.0% of females)30 and in Canadian Indians (41.6%)31 and support the report of a 60 per cent prevalence of the MetS in a sample of South African Indians with MI6, indicating a high cardiovascular risk in this community.
We considered other possibilities to explain the negative findings in our study. It was unlikely that our sample was skewed since all but LPL HinfI were in HWE. The deviation for the LPL HinfI in our study could be explained by one or more assumptions. For example, population stratification and non-random mating may be possible. Finally, genotyping errors, i.e. ‘null alleles’ being present resulting in false observation of homozygotes, could account for deviation from HWE. Furthermore, sequencing was performed in five per cent of samples to ensure the accuracy of our findings.
In our sample, females had a higher prevalence of the MetS compared to males. Although we had fewer males in our study, our finding could be true since the female gender predisposition was also shown in one other South African study6. In contrast, Chow et al32 in Southern India showed that males were at a greater risk for the MetS than females (26.9 vs 18.4%). While these gender differences could be due to the different gender ethnic-specific cut-offs in the criteria for the MetS, environmental changes and sedentary lifestyles could also have contributed to the cardiometabolic risk factor profiles and the observed gender differences in prevalence of the MetS.
Central obesity (waist circumference) (95.0%) and hypertriglyceridaemia (71.1%) were found to occur more frequently in our participants. This has been attributed to the effects of high blood glucose on lipid metabolism, resulting in hypertriglyceridaemia and obesity. The high prevalence of increased triglyceride and fasting blood glucose with the MetS suggests that risk factor clustering in our participants may be related more to obesity and diabetes than to a genetic predisposition to the MetS.
Although there was no association with the selected gene SNPs with the MetS, certain genotypes were associated with specific risk factor components of the MetS. Males diagnosed with MetS with the human paraoxonase 1 192Arg/Gln (AA genotype) were more inclined to have elevated systolic BP. It has been postulated that participants with this genotype who have hypertension may possibly have increased oxidative stress that alters the functioning of paraoxonase33, leading to endothelial dysfunction and predisposition to the MetS. More studies in this population are required to further examine the association of this SNP with MetS.
The adiponectin 45T>G SNP has been shown to be positively associated with the MetS in 151 Uygur Asians34. Although no such association was found in our study, it was observed that males diagnosed with MetS with the adiponectin 45T>G were more inclined to have reduced HDL-C levels. Larger studies are required to confirm whether the adiponectin 45T>G confers a greater risk for dyslipidaemia in male participants, in this way predisposing them to the MetS.
There are several limitations of this study that need to be considered. First, the discrepancies in associations between the SNPs and the MetS may possibly be related to the sample studied which comprised a larger number of females. This investigation used cross-sectional data, which provided information on a once-off basis and were limited by the lack of a longitudinal analysis which could contribute immensely to the understanding of the MetS and its association with genetic factors in Asian Indians. Second, the study examined specific SNPs related to lipid metabolism and to obesity; we did not genotype other SNPs known to be associated with lipid metabolism (such as FABP2 Ala54Thr20; APOA1 T655C, T756C, T1001C22), but the selected SNPs were chosen on the basis of previous studies4101719. Because of the high prevalence of diabetes in this sample, SNPs in genes associated with MetS that affect glucose metabolism such as TCF7L235 and PPAR-Gamma2324 are likely to yield more positive associations with the MetS. Third, there was a lack of haplotype analysis in this study and replicating these findings and testing for haplotypes associated with the risk for the MetS might prove valuable. The sample size for interaction-type analysis between the single locus did not reveal any significant findings which may be partially explained by the genotype/allele frequencies of the participants studied.
In conclusion, our study showed a high prevalence of the MetS in the studied population, however, no genetic predisposition to the MetS based on the studied SNPs was demonstrated. This suggests that the absolute genetic risk for the MetS is probably small and possibly lies in the component risk factors of the MetS. In view of the association with risk factor components, the adiponectin 45T>G and the human paraoxonase 1 192Arg/Gln SNPs in male participants may predispose to dyslipidaemia and hypertension, respectively. Future studies addressing gene-environmental influences are more likely to identify predisposition to the MetS.
Financial support & sponsorship: The research was financially supported by the WHO, Pfizer Laboratory (SA), and the South African Centre for Epidemiological Modelling and Analysis (SACEMA), South Africa.
Conflicts of Interest: None.
References
- The metabolic syndrome in South Asians: Epidemiology, determinants, and prevention. Metab Syndr Relat Disord. 2009;7:497-514.
- [Google Scholar]
- Linkage of the metabolic syndrome to 1q23-q31 in Hispanic families: The insulin resistance atherosclerosis study family study. Diabetes. 2004;53:1170-4.
- [Google Scholar]
- Apolipoprotein A-V gene polymorphisms in subjects with metabolic syndrome. Clin Chem Lab Med. 2007;45:1133-9.
- [Google Scholar]
- Obesity-associated genetic variants in young Asian Indians with the metabolic syndrome and myocardial infarction. Cardiovasc J Afr. 2011;22:25-30.
- [Google Scholar]
- High prevalence of cardiovascular risk factors in Durban South African Indians: The Phoenix lifestyle project. S Afr Med J. 2016;106:284-9.
- [Google Scholar]
- WHO STEPwise approach to surveillance instrument v.1.4. Geneva: World Health Organization; 2003.
- [Google Scholar]
- Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640-5.
- [Google Scholar]
- Apoa5 Q139X truncation predisposes to late-onset hyperchylomicronemia due to lipoprotein lipase impairment. J Clin Invest. 2005;115:2862-9.
- [Google Scholar]
- Complete rescue of lipoprotein lipase-deficient mice by somatic gene transfer of the naturally occurring LPLS447X beneficial mutation. Arterioscler Thromb Vasc Biol. 2005;25:2143-50.
- [Google Scholar]
- DNA variants at the LPL gene locus associate with angiographically defined severity of atherosclerosis and serum lipoprotein levels in a Welsh population. Arterioscler Thromb. 1994;14:1090-7.
- [Google Scholar]
- Effects of lipoprotein lipase gene variations, a high-carbohydrate low-fat diet, and gender on serum lipid profiles in healthy Chinese Han youth. Biosci Trends. 2011;5:198-204.
- [Google Scholar]
- Cardiovascular diseases in Chinese, Malays, and Indians in Singapore. II. Differences in risk factor levels. J Epidemiol Community Health. 1990;44:29-35.
- [Google Scholar]
- Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265-76.
- [Google Scholar]
- CETP gene variation: Relation to lipid parameters and cardiovascular risk. Curr Opin Lipidol. 2004;15:393-8.
- [Google Scholar]
- Genetic association study of adiponectin polymorphisms with risk of type 2 diabetes mellitus in Korean population. Diabet Med. 2005;22:569-75.
- [Google Scholar]
- Prevalence of obesity in a rural Asian Indian (Bangladeshi) population and its determinants. BMC Public Health. 2015;15:860.
- [Google Scholar]
- Identification of leptin gene variants in school children with early onset obesity. J Med Assoc Thai. 2009;92(Suppl 7):S108-14.
- [Google Scholar]
- Polymorphisms in the fatty acid-binding protein 2 and apolipoprotein C-III genes are associated with the metabolic syndrome and dyslipidemia in a South Indian population. J Clin Endocrinol Metab. 2005;90:1705-11.
- [Google Scholar]
- APOC3 promoter polymorphisms C-482T and T-455C are associated with the metabolic syndrome. Arch Med Res. 2007;38:444-51.
- [Google Scholar]
- Apo lipoprotein A1 gene polymorphisms predict cardio-metabolic risk in South Asian immigrants. Dis Markers. 2012;32:9-19.
- [Google Scholar]
- Genetic variants associated with insulin resistance and metabolic syndrome in young Asian Indians with myocardial infarction. Metab Syndr Relat Disord. 2008;6:209-14.
- [Google Scholar]
- Absence of association of metabolic syndrome with PPARGC1A, PPARG and UCP1 gene polymorphisms in Asian Indians. Metab Syndr Relat Disord. 2007;5:153-62.
- [Google Scholar]
- Association of Taq 1B CETP polymorphism with insulin and HOMA levels in the population of the Canary Islands. Nutr Metab Cardiovasc Dis. 2011;21:18-24.
- [Google Scholar]
- Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med (Berl). 2003;81:428-34.
- [Google Scholar]
- Linkage and association studies of the lipoprotein lipase gene with postheparin plasma lipase activities, body fat, and plasma lipid and lipoprotein concentrations: The HERITAGE family study. Metabolism. 2000;49:432-9.
- [Google Scholar]
- Can we apply the national cholesterol education program adult treatment panel definition of the metabolic syndrome to Asians? Diabetes Care. 2004;27:1182-6.
- [Google Scholar]
- Comparison of metabolic syndrome definitions in the prediction of diabetes over 5 years in Mauritius. Diabetologia. 2003;46:A145-6.
- [Google Scholar]
- A comparison of the new international diabetes federation definition of metabolic syndrome to WHO and NCEP definitions in Chinese, European and South Asian origin adults. Ethn Dis. 2007;17:522-8.
- [Google Scholar]
- Relationship of metabolic syndrome and fibrinolytic dysfunction to cardiovascular disease. Circulation. 2003;108:420-5.
- [Google Scholar]
- Significant lipid, adiposity and metabolic abnormalities amongst 4535 Indians from a developing region of rural Andhra Pradesh. Atherosclerosis. 2008;196:943-52.
- [Google Scholar]
- The influence of PON1 192 polymorphism on endothelial function in diabetic subjects with or without hypertension. Hypertens Res. 2008;31:507-13.
- [Google Scholar]
- Associations between 45T/G polymorphism of the adiponectin gene and plasma adiponectin levels with type 2 diabetes. Clin Exp Pharmacol Physiol. 2007;34:1287-90.
- [Google Scholar]
- Association of TCF7L2 polymorphism with diabetes mellitus, metabolic syndrome, and markers of beta cell function and insulin resistance in a population-based sample of Emirati subjects. Diabetes Res Clin Pract. 2008;80:392-8.
- [Google Scholar]






