Translate this page into:
Direct identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) from positive blood culture bottles: An opportunity to customize growth conditions for fastidious organisms causing bloodstream infections
Reprint requests: Dr Pallab Ray, Department of Medical Microbiology, Postgraduate Institute of Medical Education & Research, Chandigarh 160 012, India e-mail: drpallabray@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Culture-negative bacteraemia has been an enigmatic entity with respect to its aetiological agents. In an attempt to actively identify those positive blood cultures that escape isolation and detection on routine workflow, an additional step of MALDI-TOF MS (matrix-assisted laser desorption ionization-time of flight mass spectrometry) based detection was carried out directly from the flagged blood culture bottles. Blood samples from 200 blood culture bottles that beeped positive with automated (BACTEC) system and showed no growth of organism on routine culture media, were subjected to analysis by MALDI-TOF MS. Forty seven of the 200 (23.5%) bacterial aetiology could be established by bottle-based method. Based on these results, growth on culture medium could be achieved for the isolates by providing special growth conditions to the fastidious organisms. Direct identification by MALDI-TOF MS from BACTEC-positive bottles provided an opportunity to isolate those fastidious organisms that failed to grow on routine culture medium by providing them with necessary alterations in growth environment.
Keywords
Culture-negative bacteraemia
fastidious organisms
MALDI-TOF MS
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI- TOF MS) has revolutionized microbial identification from conventional biochemical-based methods to avant-garde protein profiling of individual organisms. To obviate the delay in culturing organisms, identification directly from clinical specimens has been tried12. One important specimen for which mortality increases by 7.6 per cent for every hour of delay in initiation of therapy is blood culture3. Although several groups have harnessed MALDI-TOF MS for direct identification from positive blood culture bottles45, the issue of culture-negative bacteraemia, especially with fastidious organisms has largely remained neglected till now. The aim of this study was to find out if direct identification of organism from positive blood culture bottles by MALDI-TOF MS could serve as a guide for providing modified growth conditions to uncommon fastidious organisms that had remained unrecognized due to failure to grow on ordinary medium under routine aerobic incubation.
As a routine workflow in the microbiology laboratory of the department of Medical Microbiology, Postgraduate Institute of Medical Education and Research, Chandigarh, India, all blood culture bottles are continuously monitored automatically inside BACTEC 9240 (BD Becton Dickinson, USA), Gram stain smears are prepared from beep-positive bottles and sub-cultured onto five per cent sheep blood agar and MacConkey agar. All differentiated colonies obtained after overnight incubation at 37°C are identified by MALDI-TOF MS (Microflex LT, Bruker Daltoniks, GmbH, Germany) using Biotyper 3.0 (Bruker Daltoniks).
Following this workflow, it was noticed that there were some positive bottles that failed to grow organisms on routine subculture. Between September 2013 and September 2015, of the 74,482 blood culture bottles received in the department of Microbiology, 7389 (9.92%) were true positive (i.e. beep positive and culture positive) and 66,893 were true negative (i.e. no beep till the set protocol of five days of incubation inside BACTEC machine). There were 200 bottles that were neither true positive nor true negative. These were beep positive but culture-negative bottles that were being ‘missed’ on routine workflow. To find out at what step of processing these isolates were being missed, an additional step of MALDI-identification directly from the BACTEC bottles was introduced. Blood (2 ml) withdrawn from these blood culture bottles was centrifuged at 150 g for 10 min. A volume of one ml of its supernatant was centrifuged at 13000 g for five minutes. The pellet was washed thrice with distilled water. After removing excess water, about 20 μl of pellet was achieved, one μl of which was then inoculated onto the MALDI plate. It was overlaid with 0.5 μl formic acid and one μl of matrix solution (α-cyano-4-hydroxycinnamic acid) sequentially. Once dried, the plate was inserted into the MALDI-TOF MS machine for microbial identification. Following manufacturer's instructions, a score >2 was taken as cut-off for reliable identification. Based on the identification results, these isolates were subcultured from the original bottles under modified growth conditions and the colonies so obtained were re-identified by MALDI-TOF MS.
Forty seven of the 200 (23.5%) ‘routinely missed’ isolates could be identified by bottle-based method. Using this identification for customizing culture conditions, growth could be achieved in all. Thus, the step of providing optimal growth conditions to fastidious organisms as indicated by direct MALDI-TOF MS identification from flagged bottles served as the missing link for these cases of culture-negative bacteraemia. The Table presents the details of this altered workflow.
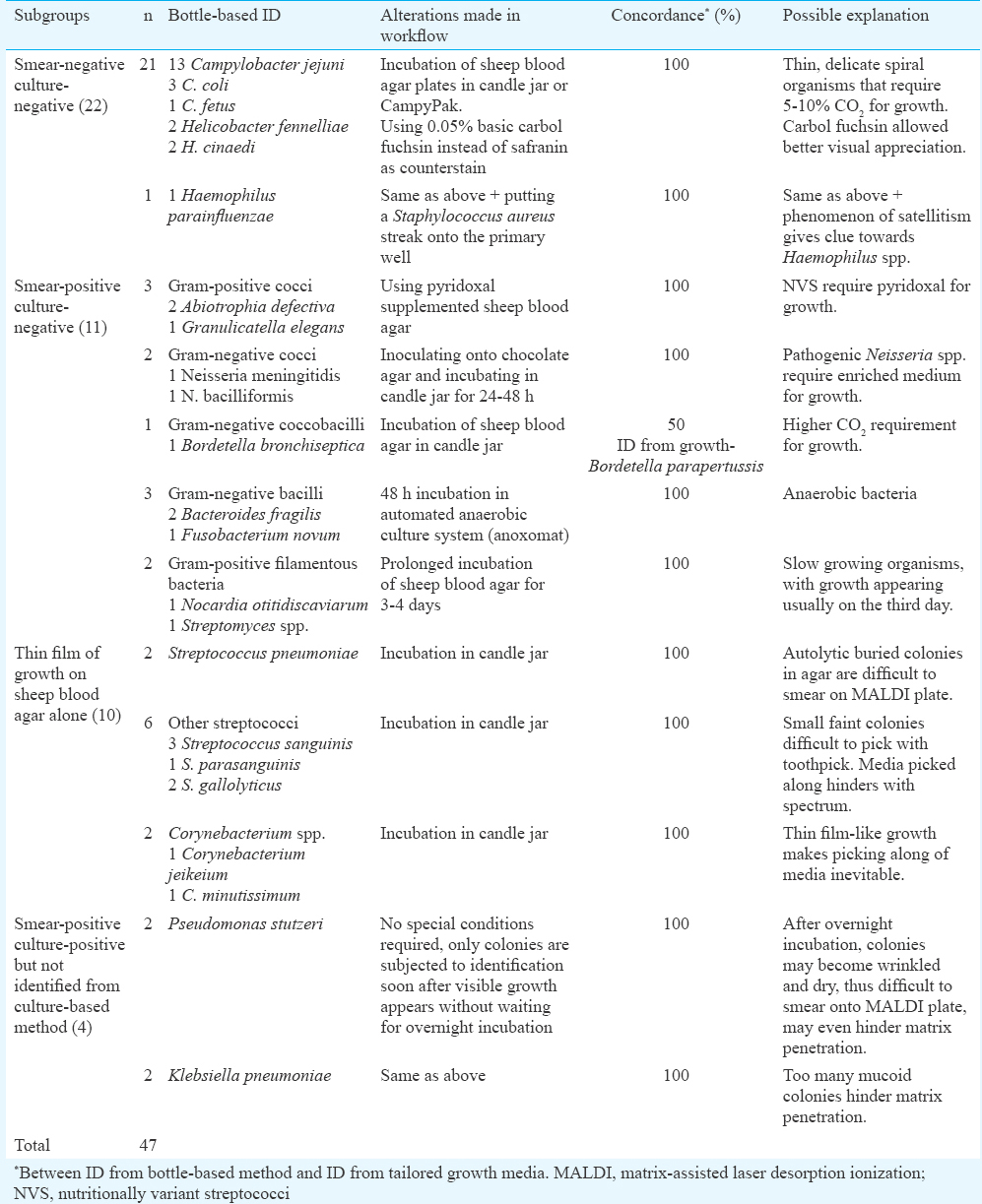
Multicentre studies have shown that culture-negative bacteraemia accounts for 30 to 49 per cent of all cases of severe sepsis67. Lack of microbiological culture evidence contributes to diagnostic dilemma thus delaying institution of appropriate therapy. Considering the high prevalence and the increased mortality, hospital stay and cost associated with culture-negative sepsis8, it would be prudent to be on an active look-out for those fastidious organisms that can be conveniently cultured by a few modifications in the routine workflow.
In our study 153 of 200 (76.5%) isolates still remained beep-positive yet unidentified by MALDI-TOF MS. Several factors could have contributed to this observation. First, low bacterial load of the fastidious organism in the inoculum could be the reason, although almost similar inoculum size of 5×108 cfu/ml was required for BACTEC bottle to beep positive and MALDI- TOF MS to identify organism directly from bottle9, the fact that only one μl of the washed pellet was smeared onto the MALDI plate explained non-identification of those isolates that had an inoculum size just near the threshold limit of BACTEC which was inadvertently reduced 20-fold during the steps of processing. Using larger initial volumes of blood from the blood culture bottles, as previously done by Rodríguez-Sánchez et al5, might reduce such procedural flaw. Another reason for low inoculum could be exposure to antibiotic treatment before blood sampling. Resin containing BACTEC bottles may provide some protection from such residual antibiotic effect. Second, the relatively crude method of extracting the organism might have caused peaks from broth remnants to interfere with the spectra. Other groups have documented higher rates of identification (59-99%) using commercially available kits and chemical lysis buffers41011. Third, the higher score cut-off of 2 might have been responsible for low identification yield. Prior studies have reported higher identification rates using score cut-off values of 1.5 and as low as 1.111. These studies, however, made use of commercial kits to minimize background peaks. Since no such specialized extraction procedure was used in our study, it was thought necessary to comply with the manufacturer recommended cut-off of 2. Fourth, false positive beep due to non-infectious causes such as leukaemias, documented between one12 and 10 per cent13, could have caused culture-negativity. Fifth, a polymicrobial infection could have interfered with the characteristic mass spectra of either of the organisms thus producing non-reliable results. This factor, however, must have made a small contribution if at all because though 10-20 per cent of all bloodstream infections can be polymicrobial14, it is highly unlikely that all causative organisms would be fastidious enough to grow on ordinary culture medium. Finally, theoretical possibility exists that the spectrum of the organism was absent from MALDI-TOF MS's database. As the database gets upgraded, exact contribution by such organisms in culture-negative bacteraemia will be revealed.
The study highlights how a simple additional step in the routine workflow of a microbiology laboratory can help in identifying those fastidious organisms that have so far remained hidden from detection despite deteriorating patient's condition. Harnessing the potential of MALDI-TOF for protein-based, culture-independent identification from a large number of organisms present in its database can go a long way in making a prompt diagnosis for septicaemia patients. The in-house protocol advocated in this study not only cut down any extra cost for a high throughput setting but also did not compromise on the yield of detection. A possible limitation of the study was that only 23.5 per cent (47/200) of the ‘routinely missed’ isolates could be detected when direct identification using MALDI-TOF was employed.
To conclude, the BACTEC-MALDI-TOF combination can be utilized to identify those fastidious organisms that have been contributing to a considerable number of culture-negative bacteraemia cases. Reliable and cost-effective identification within an hour of positive beep, followed by tailoring growth conditions will go a long way in debarring many fastidious organisms from their egregious tag of ‘causes of culture-negative bacteraemia’.
Conflicts of Interest: None.
References
- Direct application of MALDI-TOF mass spectrometry to cerebrospinal fluid for rapid pathogen identification in a patient with bacterial meningitis. Clin Chim Acta. 2014;435:59-61.
- [Google Scholar]
- Rapid urine preparation prior to identification of uropathogens by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015;34:1787-95.
- [Google Scholar]
- Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589-96.
- [Google Scholar]
- Identification of pathogenic microorganisms directly from positive blood vials by matrix-assisted laser desorption/ionization time of flight mass spectrometry. APMIS. 2013;121:871-7.
- [Google Scholar]
- Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2014;20:O421-7.
- [Google Scholar]
- International study of the prevalence and outcomes of infection in Intensive Care Units. JAMA. 2009;302:2323-9.
- [Google Scholar]
- The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546-54.
- [Google Scholar]
- Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit Care. 2013;17:R202.
- [Google Scholar]
- Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48:1584-91.
- [Google Scholar]
- Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol. 2013;51:1733-9.
- [Google Scholar]
- Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J Mol Diagn. 2011;13:701-6.
- [Google Scholar]
- Direct identification of bacteria from positive blood cultures by amplification and sequencing of the 16S rRNA gene: Evaluation of BACTEC 9240 instrument true-positive and false-positive results. J Clin Microbiol. 2001;39:3578-82.
- [Google Scholar]
- PCR evaluation of false-positive signals from two automated blood-culture systems. J Med Microbiol. 2006;55:53-7.
- [Google Scholar]
- Population-based assessment of the incidence, risk factors, and outcomes of anaerobic bloodstream infections. Infection. 2013;41:41-8.
- [Google Scholar]






