Translate this page into:
Comparative analysis of inflammatory gene expression levels in metabolic syndrome & coronary artery disease
Reprint requests: Dr. Jayashree Shanker, Mary & Garry Weston Functional Genomics Unit, Thrombosis Research Institute, Narayana Hrudayalaya 258/A, Bommasandra Industrial Area, Anekal Taluk, Bengaluru 560 099, Karnataka, India e-mail: jayashreeshanker@triindia.org.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Metabolic syndrome (MetS) increases the likelihood of developing coronary artery disease (CAD), and inflammation is involved in the pathogenesis of both these conditions. The present work was conducted to examine the relative expression of 18 key inflammatory genes associated with MetS and incident CAD in a representative group of patients.
Methods:
A total of 178 male patients, including 57 with CAD and 121 without CAD, were enrolled in the study. The participants without CAD were characterized for the presence of MetS using modified criteria specific for Asian Indians, which included a lower cut-off for waist circumference (≥90 cm for men). The expression of 18 inflammatory genes was evaluated in peripheral whole blood by quantitative polymerase chain reaction method.
Results:
Of the 121 participants without CAD, 53 (43.8%) had three or more risk factors (MetS group), 50 (41.3%) had one or two risk factors (non-MetS group), while 18 (14.8%) did not have any risk factors (control group). High nuclear factor-kappa B (NF-κB) expression levels and low interleukin-10 (IL-10) levels were observed in MetS patients. Linear association was seen between NF-κB and vascular endothelial growth factor A (VEGFA) expression and with increase in MetS components. Comparison of gene expression pattern between CAD and MetS revealed significantly higher expression of leukotriene genes - arachidonate 5-lipoxygenase (ALOX5), arachidonate 5-lipoxygenase activating protein (ALOX5 AP), leukotriene A4 hydrolase (LTA4H) and leukotriene C4 synthase (LTC4S), and lower expression of NF-κB, interleukin 1 beta (IL-1β), monocyte chemoattractant protein-1 (MCP-1/CCL2) and signal transducer and activator of transcription 3 (STAT3) genes in CAD. There was linear increase in expression of LTA4H, LTC4S, IL-8 and VEGFA genes across the four groups, namely from controls, non-MetS, MetS and CAD.
Interpretation & conclusions:
A distinct gene expression pattern was seen in MetS and CAD implying a well-orchestrated inflammatory and immune activity. Specifically, NF-κB might be playing an active role in MetS, allowing further expansion of the inflammatory process with resolution of inflammation in full-blown CAD, wherein other gene players such as leukotrienes may dominate.
Keywords
Coronary artery disease
gene expression
inflammation
metabolic syndrome
Extensive research support the prominent role of inflammation in the progression and manifestation of coronary artery disease (CAD)1. The pivotal role of inflammatory cells, inflammatory proteins and inflammatory response across the different stages of atherosclerosis, including initiation, progression and plaque rupture, is well known1. It has been postulated that induction of this inflammatory process can lead to chronic complications such as hypertension, dyslipidaemia, insulin resistance, diabetes mellitus, metabolic syndrome (MetS) and atherosclerosis2. While the underlining cause of MetS is debatable, an imbalance between increased inflammatory stimuli and decreased anti-inflammatory mechanisms could be a possible working hypothesis3. Gene expression profiling studies carried out on MetS, CAD, type 2 diabetes mellitus (T2DM) and rheumatoid arthritis provide strong evidence for the role of inflammation in the pathogenesis of these diseases with unique immune processes being involved4. Sarkozy et al5 have shown that MetS can alter the cardiac gene expression pattern in male Zucker diabetic fatty rats. These studies indicated that inflammation could be an important element linking MetS and CAD. Although it is known that the presence of MetS is associated with increased cardiovascular risk, the extent and association of specific inflammatory markers have not been fully enumerated. This study was aimed at understanding the contribution of select inflammatory markers in patients with MetS and CAD. To address this, the expression of a select panel of 18 inflammatory genes was studied in patients with MetS and CAD and the association and correlation patterns of relative gene expression were compared with each other and against controls. These 18 genes were selected based on bioinformatics analysis of the inflammatory bionetwork previously carried out for gene prioritization6 and through a selection of highly networked leukotriene-induced inflammatory genes described elsewhere7.
Material & Methods
A total of 178 male participants, comprising 57 with symptomatic CAD and 121 age-matched male individuals without CAD, were included in this study. These were enrolled during May 2012 to February 2014, and were randomly selected from Indian Atherosclerosis Research Study (IARS)8. The IARS is an ongoing epidemiological study with an aim to investigate the genetic factors and biomarkers associated with CAD along with their risk factors in the Asian Indian population. The IARS families were enrolled from Narayana Institute of Cardiac Sciences and other local hospitals/clinics in Bengaluru, in southern India, and the Asian Heart Institute and Research Centre, Mumbai, in Western India. A detailed design of the IARS is published elsewhere89. Given a sample size of 178, and considering the effect size to be medium, β/α ratio of 4, with a standard value of 0.20 and 0.05 for β and α, respectively, the power of study was estimated to be 95 per cent (G*Power 3.1.6 tool, (Franz Faul, University of Kiel, Kiel, Germany).
Symptomatic individuals with CAD were defined as those with abnormal electrocardiogram (ECG), angiographically confirmed the presence of CAD, having >70 per cent stenosis in any one major epicardial artery or >50 per cent stenosis in two or more arteries, those undergoing percutaneous transluminal coronary angioplasty or bypass graft surgery. This included 63 per cent chronic stable angina patients and 37 per cent myocardial infarction cases. CAD severity was defined based on the number of diseased coronary vessels as one, two or three or more, showing >50 per cent luminal narrowing of the vessels. Control subjects were clinically asymptomatic for CAD, having no known history of CAD or sudden death in the family and normal ECG and were characterized for the presence of MetS. All participants were free from concomitant infection (HIV, hepatitis B surface antigen, hepatitis C virus, cold, cough, fever) and other major illnesses (cancer, liver failure, arthritis, etc.). All participants selected for the study provided informed written consent. The study was approved by the institutional ethics committee.
Definition and classification of metabolic syndrome (MetS): The presence of MetS in this study participants was determined based on the modified criteria suitable for Asian Indians9, which includes lower cut-offs for waist circumference (WC) ≥90 cm, serum triglyceride (TG) ≥150 mg/dl (1.7 mmol/l), high-density lipoprotein-cholesterol (HDL-C) ≤40 mg/dl (1.03 mmol/l), blood pressure of ≥130/85 mmHg and fasting blood glucose (FBG) level of ≥110 mg/dl (6.1 mmol/l). Individuals having three or more of the five risk components were classified as having MetS. A MetS score of 0-5 was determined based on the number of MetS components present in an individual.
The 121 individuals without CAD were characterized for the presence of MetS. Individuals having three or more risk factors were categorized into MetS, those with one or two risk factors into non-MetS group while individuals having no risk factors served as controls. A score of 0 to >4 was assigned based on the number of MetS components present in an individual. Group-wise breakdown of the 121 individuals was as follows: (i) 18 without any risk factors (control group), (ii) 50 with one or two risk factors (non-MetS group), and (iii) 53 with three or more risk factors (MetS group) (Table I). In most groups, abdominal obesity was the most common feature, followed by low HDL-C and high TG levels.
Data collection: Detailed demographics, anthropometrics, vital parameters, medical history, medication and pedigree information were recorded for each participant. Presence of T2DM, hypertension and CVD was ascertained based on self-report of physician's diagnosis and/or use of prescription medication along with perusal of their medical records. Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were measured. BMI was calculated as a ratio of weight in kilogram to height in square metre.
Laboratory assay: Venous blood (22 ml) was collected in evacuated tubes after overnight fasting of 10-12 h. Serum, EDTA and citrate plasma samples were separated by centrifugation within two hours of sampling and aliquots were stored at −80°C for biochemical analysis. Plasma levels of total cholesterol (TC), TG, HDL-C and FBG were measured using reagents from Siemens Dimension Flex reagent cartridge (Siemens Healthcare Diagnostics Ltd., UK) and standards from Randox laboratories (Crumlin, UK) on Siemens dimension Xpand plus instrument (Siemens Dade Behring, Liederbach, Germany). Liquichek lipid control was used as an assayed quality control (Bio-Rad, USA) that was run along with every experiment. Low-density lipoprotein-cholesterol (LDL-C) was calculated by applying Friedewald's formula10.
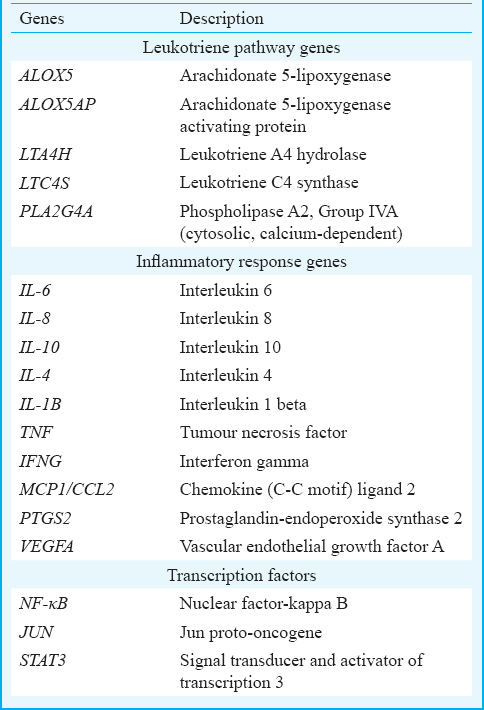
Selection of candidate genes: The network approach helped identify key inflammatory genes from a large panel of genes involved in the inflammatory process and the direct interactions among the genes in the leukotriene pathway67. The 18 genes prioritized for the present study are listed in Table II and include the leukotriene pathway genes, inflammatory genes and key transcription factors.
Real-time quantitative PCR (qPCR) assay: Total RNA was isolated from 3 ml of fresh whole blood sample using QIAamp RNA Blood Mini Kit (Qiagen Inc., Germany), treated with DNase I (Fermentas, Canada) and reverse transcribed to cDNA using cDNA archive kit (Applied Biosystems, USA), following the manufacturer's instructions. RNA was quantified by absorbance at 260 nm and the purity was estimated by the ratio A260/A280 nm and A260/A230 nm using NanoDrop 1000 TM (Thermo Fisher Scientific, USA). Quantitative real-time polymerase chain reaction (q-PCR) was performed in duplicates on 7900 HT Fast RT-PCR system (Applied Biosystems) either based on TaqMan or SYBR green chemistry. Arachidonate 5-lipoxygenase (ALOX5), arachidonate 5-lipoxygenase activating protein (ALOX5AP) and leukotriene A4 hydrolase (LTA4H) gene expression was measured using TaqMan chemistry and the primer/probe pairs with the following assay IDs - Hs01095330_m1 (ALOX5), Hs00233463_m1 (ALOX5AP) and Hs1075882_ml (LTA4H), were purchased from Applied Biosystems.
The primer pairs used for the SYBR green-based gene expression assay were selected either from PrimerBank11 or designed using PrimerQuest® program (Integrated DNA Technologies, Coralville, USA) for those genes where the primers were not available in PrimerBank. All primer sequences were further verified using BLAST search. The RT-PCR efficiency of each primer pair was determined by constructing the standard curve with serial dilution of sample. Relative quantitation for the Taqman assay was calculated using RQ Manager v1.2 software (Applied Biosystems) while the relative expression for SYBR green assay was estimated by comparative Ct method12. Beta-glucuronidase (GUSB) gene served as the endogenous control. The expression levels were initially normalized to an endogenous control and mRNA abundance in each sample was determined in relation to a reference sample (calibrator). All assays were set up in duplicates. The outlier samples were re-tested and persistent outliers were excluded from further analysis.
Statistical analysis: Student's t test and multivariate analysis were used to determine the differences in normalized mRNA expression levels and other quantitative traits between the cases and controls. Normality distribution of mRNA expression levels was assessed with Q-Q plot. Statistical differences for experiments with three or more groups were determined using analysis of variance (ANOVA), and the linearity of association was tested with polynomial contrast for linear trend. Binary logistic regression analysis was used to assess the association of mRNA expression with CAD and to estimate the corresponding odds ratio (OR). Pearson's correlation was performed to evaluate the correlation in gene expression between the genes under study. Age, hypertension, diabetes and WC was considered as potential confounders and appropriately adjusted during analysis. All analyses were performed using SPSS v 17.0 (SPSS Inc., Chicago, IL, USA) statistical software package. Quantitative variables are expressed as mean±standard error of mean, unless stated otherwise. All statistical tests were two-sided, with 95 per cent confidence interval (CI).
Results
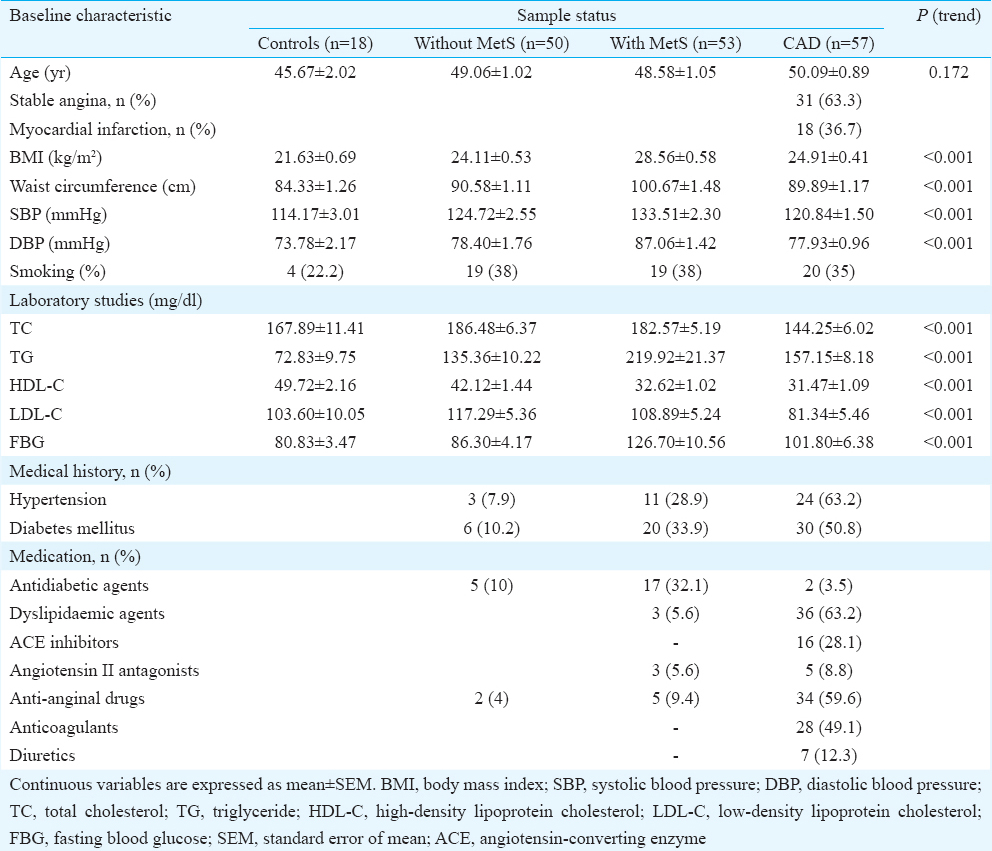
The clinical and biochemical characteristics of the study participants with and without MetS and with CAD are presented in Table III. The mean age ranged from 45 to 50 yr in controls, MetS cases and CAD. CAD patients had low TC and LDL-C levels, which might be attributed to the usage of lipid lowering drugs in this group. As anticipated, the usage of prescription drugs were high in CAD patients as more than 60 per cent were on lipid lowering drug such as statins and about 60 per cent were on anti-anginal drug such as beta-blockers. A significant difference in the SBP, DBP, WC and BMI was observed in MetS subjects as compared to the non-MetS group.
Checking for confounders: The association of inflammatory gene expression with the common confounding factors such as age, smoking, diabetes, hypertension, WC and statin was assessed using multivariate analysis. This analysis showed that the expression levels of nuclear factor-kappa B (NF-κB) and vascular endothelial growth factor A (VEGFA) were associated with age, diabetes, hypertension and WC, and hence, these factors were considered as confounders and appropriately adjusted for during analysis.
Metabolic syndrome and inflammatory gene expression: Analysis of individuals with (n=53) and without (n=50) MetS showed that there was higher expression of NF-κB (0.69±0.07 vs. 0.33±0.04, P<0.001) and lower expression of interleukin-10 (IL-10) (1.74±0.15 vs. 3.37±0.58, P=0.007) in the presence of MetS (Fig. 1). The significance was retained even after adjusting for age, diabetes, hypertension and WC. Furthermore, there was significant association of NF-κB with MetS when the top NF-κB quartile was compared to the bottom NF-κB quartile (OR=7.72, 95% CI 2.15-27.72, P= 0.002).
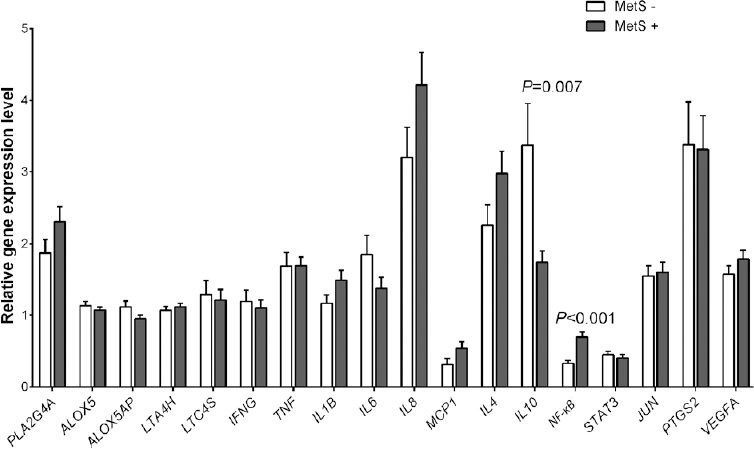
- Mean expression levels of inflammatory markers in individuals with and without metabolic syndrome (MetS). Data are represented as mean±standard error. The full forms of the genes are listed in the Table II.
When individuals with MetS (n=53) were compared with the control group having no risk factors (n=18), there was significant higher expression of LTA4H (1.12±0.04 vs. 0.90±0.06, P=0.019), IL-8 (4.21±0.45 vs. 2.34±0.39, P=0.038), NF-κβ (0.69±0.07 vs. 0.36±0.05, P=0.017) and VEGFA (1.78±0.12 vs. 0.96±0.14, P=0.001) in the former group. However, following adjustment for potential confounders, significance was retained only for VEGFA.
Association based on metabolic syndrome score: There was a linear increase in NF-κB (P<0.001) and VEGFA (P= 0.012) gene expression based on the number of MetS components (0-5) present (Fig. 2). Based on MetS score, the mean expression of NF-κB and VEGFA were as follows - 0 MetS score - 0.36±0.05 and 0.96± 0.13, 1 MetS score - 0.32±0.06 and 1.4±0.14, 2 MetS score - 0.33±0.05 and 1.71±0.17, 3 MetS score - 0.61±0.11 and 1.77±0.16 and 4 or more MetS score - 0.79±0.09 and 1.79±0.2, respectively. However, significant association was not retained after adjusting for age, diabetes, hypertension and WC. Nonetheless, the linear association trend was maintained.
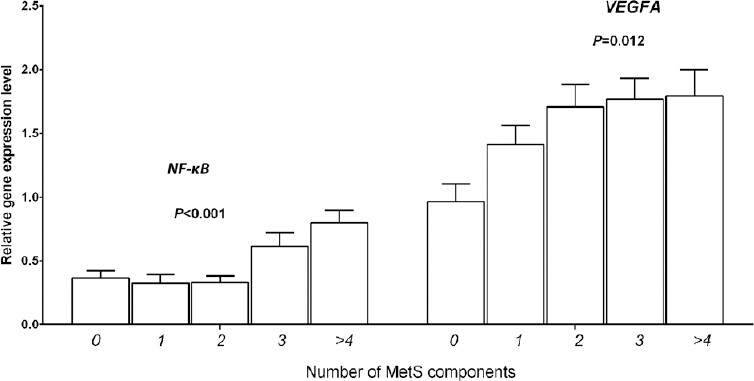
- Linear increase in nuclear factor-kappa B (NF-κB), and vascular endothelial growth factor A (VEGFA) gene expression based on the number of metabolic syndrome (MetS) components.
Comparison of gene expression pattern in MetS & CAD: Comparison of the gene expression profile between patients with MetS (n=53) and CAD (n=57) showed that there was significantly higher expression of all four leukotriene genes in CAD as compared to MetS, which included ALOX5 (1.58±0.08 vs. 1.07±0.03, P<0.001), ALOX5AP (1.40±0.07 vs. 0.95±0.04, P<0.001), LTA4H (1.31±0.06 vs. 1.12±0.04, P= 0.021), leukotriene C4 synthase (LTC4S) (2.06±0.22 vs. 1.21±0.15, P= 0.002) along with other inflammatory genes, prostaglandin-endoperoxide synthase 2 (PTGS2) (9.58±1.51 vs. 3.31±0.47, P< 0.001); IL-6 (1.94±0.24 vs. 1.38±0.15, P=0.045), IL-8 (6.73±0.68 vs. 4.21±0.45, P= 0.003), and the transcription factors, signal transducer and activator of transcription 3 (STAT3) (0.60±0.07 vs. 0.40±0.04, P= 0.021) and Jun proto-oncogene (JUN) (3.06±0.43 vs. 1.60±1.44, P=0.001). However, interferon gamma (IFNG) (0.48±0.04 vs. 1.11±0.11, P< 0.001), MCP-1 (0.13±0.02 vs. 0.54±0.09, P< 0.001 and NF-κB (0.29±0.08 vs. 0.69±0.07, P< 0.001 showed higher expression in MetS patients. Significance remained for ALOX5, ALOX5AP, LTC4S and IFNG after covariate adjustment. The distribution of mean expression levels in MetS and CAD is shown in Fig. 3.
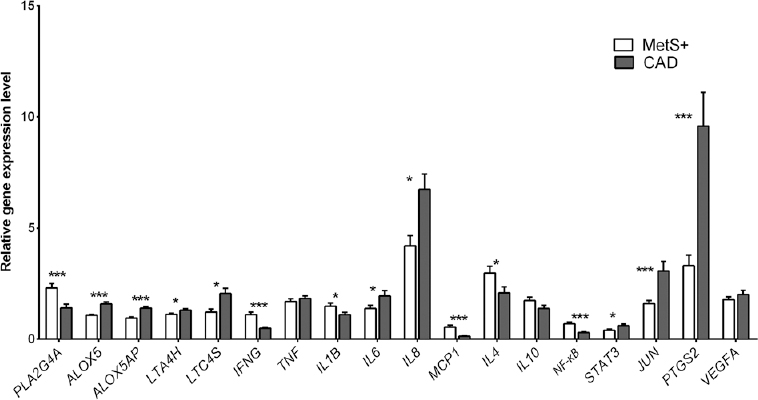
- Relative gene expression levels of inflammatory markers in patients with metabolic syndrome (MetS) and coronary artery disease (CAD). Data are represented as mean±standard error. P*<0.05, ***< 0.001. The full forms of the genes are as listed in the Table II.
Correlation analysis: There was significant correlation in the transcript expression among LTA4H, IL8, NF-κB, JUN and VEGFA genes (r=0.61-0.22) in the MetS group. A significant positive correlation was observed between the expression of NF-κB and a number of inflammatory genes - LTA4H (r=0.31), IFNG (r=0.40), TNF (r=0.38), IL1B (r=0.28), IL8 (r=0.38), MCP-1 (r=0.50), JUN (r=0.31), PTGS2 (r=0.37) and VEGFA (r=0.46). LTA4H (r=0.24, P= 0.008), IL-8 (r=0.28, P= 0.003), NF-κB (r=0.32, P= 0.001) and JUN (r=0.19, P= 0.037) correlated significantly with plasma TG levels. Furthermore, NF-κB showed correlation with WC (r=0.21, P= 0.024), BMI (r=0.26, P= 0.005), SBP (r=0.23, P= 0.12) and DBP (r=0.29, P= 0.001), while VEGFA showed significant correlation with BMI (r=0.22, P= 0.021) and FBG levels (r=0.20, P= 0.035) only.
In the CAD group, there was strong positive correlation among the leukotriene genes, ALOX5, ALOX5 AP, LTA4H and phospholipase A2, Group IVA (cytosolic, calcium-dependent) (r=0.66-0.39; P< 0.01). The anti-inflammatory gene, IL-10, showed positive correlation with ALOX5 (r=0.30), ALOX5AP (r=0.33), LTC4S (r=0.6) and NF-κB (r=0.35) transcript expression. In addition, ALOX5 showed significant correlation with WC (r=0.29) and LTA4H and LTC4S with FBG (r=0.45-0.48), while the remaining inflammatory markers did not show any significant correlation with the other MetS components.
Test for trend: Mean gene expression levels was tested for linear association trend across four groups, namely controls, non-MetS, MetS and CAD, using ANOVA, with polynomial contrast test. As shown in Fig. 4, there was linear increase of LTA4H, LTC4S, IL8, JUN, PTGS2 and VEGFA (P= 0.004-P<0.0001) across the four groups, with the highest expression levels recorded in CAD patients. In contrast, IFNG (P< 0.001) showed reduced expression in this group.
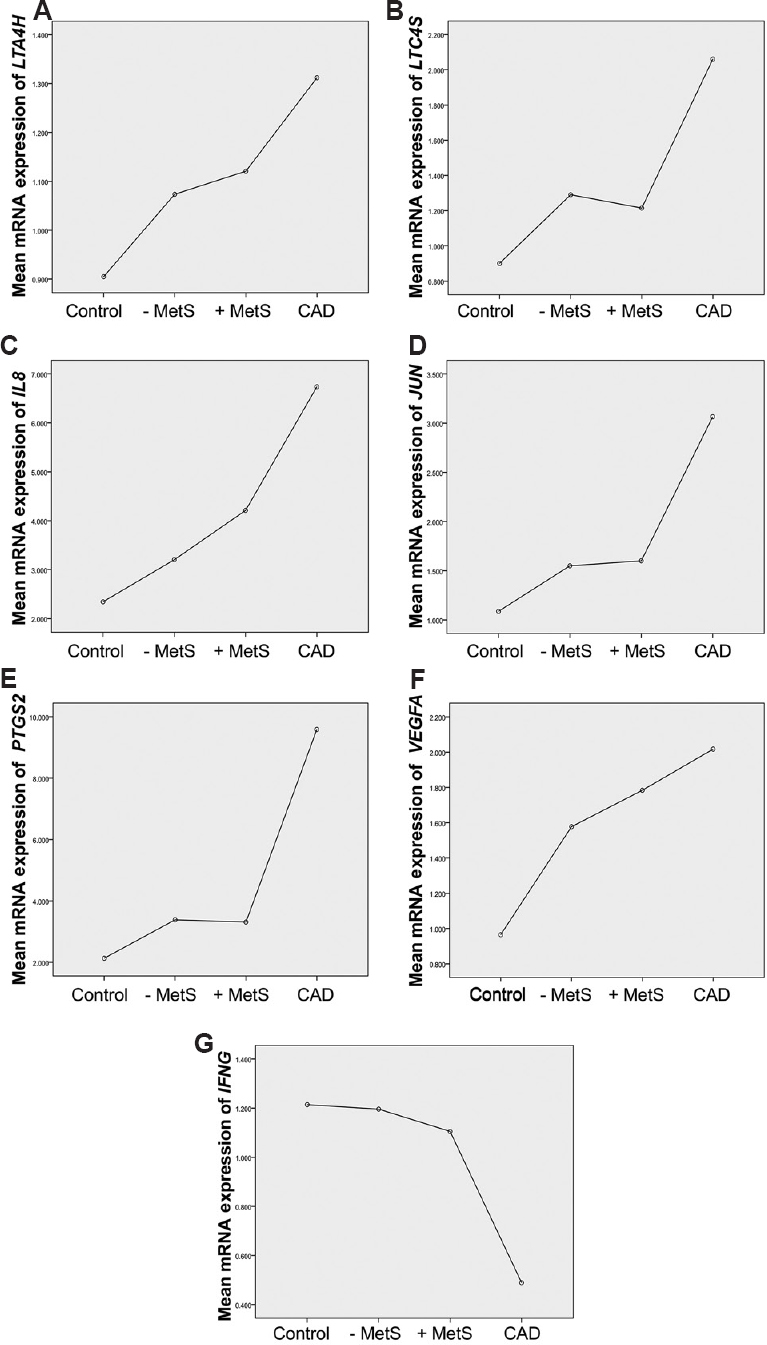
- Linear trend of inflammatory biomarkers across the different group of study participants. Mean expression of leukotriene A4 hydrolase (LTA4H) (A), leukotriene C4 synthase (LTC4S) (B), interleukin-8 (IL8) (C), Jun proto-oncogene (JUN) (D), prostaglandin-endoperoxide synthase 2 (PTGS2) (E), vascular endothelial growth factor A (VEGFA) (F) and interferon gamma (IFNG) (G) across four groups i.e. controls, without metabolic syndrome (MetS−), with MetS (MetS+) and coronary artery disease (CAD).
Discussion
A large-scale epidemiological study has shown the pleiotrophic effect of inflammatory genes with MetS13. Studies revealed inflammation to be a common pathological component in MetS and CAD but provided minimal information regarding the role of these inflammatory markers from MetS condition to full-blown CAD condition13.
In our earlier work, we have shown the predominant association of a panel of 18 inflammatory genes with CAD78. As an extension of these findings, we tested the association of these inflammatory genes and leukotriene genes with MetS and the differences if any, from incident CAD. The objective was to delineate specific genes that could be involved in the progression of MetS to its sequelae, i.e. CAD. An increased expression of NF-κB was observed only in MetS patients but not in CAD. The role of transcription factor, NF-κB, in the induction of proinflammatory gene expression is a key aspect and controlled regulation of these genes is critical for maintaining immune homoeostasis14. Kanters et al15 suggested the pivotal role of NF-κB in the induction of proinflammatory gene activity during the early onset of inflammation and the expression of anti-inflammatory genes during the resolution of inflammation. Pertaining to these observations, there was significant correlation of IL-10 with NF-κB gene expression in the CAD group only.
At present, the role of leukotrienes in the initiation and pathogenesis of MetS has not been fully elucidated. However, the importance of leukotrienes in the regulation of immune and inflammatory functions has been implicated in several inflammatory diseases including atherosclerosis16. It has been suggested that leukotrienes orchestrate the immune response by modulating the release of both pro- and anti- inflammatory cytokines17. Increased number of cells containing ALOX5 was found on the vessel wall in an advanced Stage IV CAD (a lesion that is potentially symptom-producing) as compared with Stage II and Stage III18. In our study, a higher expression of ALOX5, ALOX5AP, LTA4H and LTC4S was observed in CAD as compared to MetS patients, suggesting their predominant role in advanced disease condition. Further, reduced expression of proinflammatory genes, IL-1B, MCP-1 and increased expression of STAT3 was observed in CAD patients as compared to those with MetS19. STAT3 is activated under stress conditions19 and this activation acts as protective effect2021. Some studies have also attributed anti-inflammatory actions to STAT3 signalling in the heart through suppression of interleukin gene expression2223. Hence, the higher STAT3 and lower NF-κB expression levels and their activators such as IL-1B and MCP-1 could be possibly attributed to a regulation of inflammatory response resolution in CAD patients as compared to its precursor stage like MetS2425.
Our data showed the association of LTA4H, LTC4S, IL-8, JUN, PTGS2, VEGFA and IFNG in the progression of disease from a pre-disease to a disease state. These inflammatory genes play a key role in both MetS and CAD, but the precise role of IFNγ in atherosclerosis is not clear, having both pro- and anti-atherogenic attributes26. Several studies have shown impaired activity of IL-8 and JUN in the pathogenesis of MetS, type 2 diabetes and in atherosclerotic lesions272829. Thus, the persistence of these inflammatory molecules in the precursor stage to CAD condition underlines the activation of these molecules during early stages of disease development and their sustained aggravation up to an advanced stage.
The present study had a few limitations. First, the study included only male participants, which might have led to a gender bias to the findings. The controls were selected based on normal ECG readings. Although ECG is routinely used to rule out CAD in the absence of overt clinical symptoms, it does not preclude the presence of a silent underlying disease condition and is, therefore, not a confirmatory test. The effect of medication in the CAD group could have influenced the expression levels of inflammatory cytokines. Although no significant differential expression was observed with and without the presence of statins (data not shown), one cannot rule out the possible influence of other standard medications routinely used for treating CAD and its comorbidities. Finally, this was a cross-sectional study, and hence, it was not possible to discern between a causal and casual relationship.
In conclusion, our data suggested the activation of immune response in MetS through NF-κB and the resolution of inflammation in treated CAD condition. This suggests that NF-κB activation could be an initial trigger molecule in the slow build-up leading to plaque development. Comparison of the expression pattern of key inflammatory genes showed the pivotal role of leukotrienes in the presence of symptomatic CAD. Comparison of the expression pattern of key inflammatory genes in individuals with no-risk factors to MetS and finally CAD revealed a distinct pattern, suggesting a well-orchestrated inflammatory and immune response process at play during CAD development.
Acknowledgment
Authors acknowledge the support extended by the trustees of Thrombosis Research Institutes in London and Bengaluru, Weston Foundation, UK, Foundation Bay, Switzerland, the Tata Social Welfare Trust, India, (TSWT/IG/SNB/JP/Sdm), and the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi (BT/01/CDE/08/07).
Conflicts of Interest: None.
References
- Inflammation as a link between obesity and metabolic syndrome. J Nutr Metab. 2012;2012:476380.
- [Google Scholar]
- The metabolic syndrome and inflammation: Association or causation? Nutr Metab Cardiovasc Dis. 2004;14:228-32.
- [Google Scholar]
- Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun. 2011;12:341-51.
- [Google Scholar]
- Metabolic syndrome influences cardiac gene expression pattern at the transcript level in male ZDF rats. Cardiovasc Diabetol. 2013;12:16.
- [Google Scholar]
- Network analysis of inflammatory genes and their transcriptional regulators in coronary artery disease. PLoS One. 2014;9:e94328.
- [Google Scholar]
- Expression analysis of leukotriene-inflammatory gene interaction network in patients with coronary artery disease. J Atheroscler Thromb. 2014;21:329-45.
- [Google Scholar]
- Rationale, design & preliminary findings of the Indian Atherosclerosis Research Study. Indian Heart J. 2010;62:286-95.
- [Google Scholar]
- Prevalence and component analysis of metabolic syndrome: An Indian atherosclerosis research study perspective. Vasc Health Risk Manag. 2008;4:189-97.
- [Google Scholar]
- Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499-502.
- [Google Scholar]
- PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792-9.
- [Google Scholar]
- A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
- [Google Scholar]
- Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014;112:317-38.
- [Google Scholar]
- The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651.
- [Google Scholar]
- Inhibition of NF-kappaB activation in macrophages increases atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2003;112:1176-85.
- [Google Scholar]
- The role of leukotrienes in the pathophysiology of inflammatory disorders: Is there a case for revisiting leukotrienes as therapeutic targets? Inflammopharmacology. 2006;14:10-6.
- [Google Scholar]
- Cytokine-leukotriene receptor interactions. ScientificWorldJournal. 2007;7:1348-58.
- [Google Scholar]
- Expanding expression of the 5-lipoxygenase pathway within the arterial wall during human atherogenesis. Proc Natl Acad Sci U S A. 2003;100:1238-43.
- [Google Scholar]
- Cytokines and their receptors in cardiovascular diseases - role of gp130 signalling pathway in cardiac myocyte growth and maintenance. Int J Exp Pathol. 2000;81:1-16.
- [Google Scholar]
- Activation of signal transducer and activator of transcription 3 protects cardiomyocytes from hypoxia/reoxygenation-induced oxidative stress through the upregulation of manganese superoxide dismutase. Circulation. 2001;104:979-81.
- [Google Scholar]
- Cardiomyocyte-restricted knockout of STAT3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc Natl Acad Sci U S A. 2003;100:12929-34.
- [Google Scholar]
- Interleukin-10 protects the ischemic heart from reperfusion injury via the STAT3 pathway. Surgery. 2011;150:231-9.
- [Google Scholar]
- Therapeutic activation of signal transducer and activator of transcription 3 by interleukin-11 ameliorates cardiac fibrosis after myocardial infarction. Circulation. 2010;121:684-91.
- [Google Scholar]
- Possible new role for NF-kappaB in the resolution of inflammation. Nat Med. 2001;7:1291-7.
- [Google Scholar]
- Mechanisms of resolution of inflammation: A focus on cardiovascular disease. Arterioscler Thromb Vasc Biol. 2011;31:1001-6.
- [Google Scholar]
- Interferon gamma: A master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125-35.
- [Google Scholar]
- Association of systemic chemokine concentrations with impaired glucose tolerance and type 2 diabetes: Results from the Cooperative Health Research in the Region of Augsburg Survey S4 (KORA S4) Diabetes. 2005;54(Suppl 2):S11-7.
- [Google Scholar]
- Upregulation of interleukin-8/CXCL8 in vascular smooth muscle cells from spontaneously hypertensive rats. Hypertens Res. 2008;31:515-23.
- [Google Scholar]
- Increased expression and activation of stress-activated protein kinases/c-Jun NH(2)-terminal protein kinases in atherosclerotic lesions coincide with p53. Am J Pathol. 2000;156:1875-86.
- [Google Scholar]






