Translate this page into:
Observation on frequency & clinico-pathological significance of various cytogenetic risk groups in multiple myeloma: an experience from India
Reprint requests: Dr Pratibha S. Kadam Amare, Department of Cancer Cytogenetics, Tata Memorial Hospital, 7th Floor, Annex Bldg, Dr. Ernest Borges Road, Parel, Mumbai 400 012, Maharashtra, India e-mail: pratibha.amare@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Multiple myeloma (MM) is a plasma cell malignancy characterized by cytogenetic heterogeneity. In comparison with conventional karyotyping, fluorescence in situ hybridization (FISH) can efficiently detect various genetic changes in non-cycling plasma cells in 50-90 per cent of MM cases. The present study was undertaken in MM patients to evaluate the frequency and clinico-pathological significance of various cytogenetic abnormalities in the Indian population.
Methods:
Interphase FISH was applied on purified plasma cells of 475 patients with MM using specific probes. Interphase FISH for 1q gain/1q amplification was performed on a separate group of 250 newly diagnosed MM patients.
Results:
Low frequency of Δ13 [-13/del(13q)] (32%) and t(11;14) (5%) was observed in our 475 patients probably due to ethnic diversity. Clustering of Δ13, del(17) (p13.1) and IgH translocations in non-hyperdiploidy confirmed prognostic significance of ploidy in MM. t(4;14) and del(17) (p13.1) were high-risk groups due to correlation with high serum β2-microglobulin, increased plasma cells and advanced disease. Hyperdiploidy and t(14;16) were associated with higher age group. In a separate group of 250 patients, 1q amplification [amp(1q)] in combination with Δ13 and/or del(17p) with t(4;14) revealed association with adverse clinico-laboratory features, which confirmed progressive role of amp(1q) with adverse prognostic impact. Amp(1q) was clustered at 1q21 and 1q25 loci.
Interpretation & conclusions:
Based on our findings, it appears that comprehensive analysis of various cytogenetic aberrations by interphase FISH is a powerful strategy being adapted for risk stratification of MM.
Keywords
Δ13
1q amplification
ethnic diversity
fluorescence in situ hybridization
IgH translocations
multiple myeloma
non-hyperdiploidy
Multiple myeloma (MM) is a clonal B-cell malignancy, characterized by heterogeneity at both clinical and genomic level. Despite the role of specific cytogenetic aberrations in the pathogenesis of disease, the prognostic significance of cytogenetic changes has been identified in MM and it has become an integral part of disease management. The identification of high-risk and low-risk cytogenetic groups plays an important role in prediction of response or resistance to therapy. The cytogenetic findings are included in the consensus statement of the European Myeloma Network and International Myeloma Working Group12345678. Traditional, conventional metaphase cytogenetics has shown limitations due to low proliferative nature of the malignant plasma cells which yield poor mitotic index. Several groups have adopted interphase fluorescence in situ hybridization (FISH) which could efficiently detect genetic changes such as chromosome 13 aberrations, del(17p), various IgH (immunoglobulin heavy chain) translocations and trisomies of the odd number of chromosomes in non-cycling plasma cells in >60 per cent of MM cases1234678. Large-scale, comprehensive cytogenetic data in MM are lacking in the Indian population. The present study was designed to evaluate the frequency of cytogenetic abnormalities, to analyze their clinico-pathological correlation and to implement the information in the risk stratification of disease.
Material & Methods
A total of 475 consecutive patients (male: 345, females: 130, age range: 25-90 yr) who were diagnosed in the department of Medical Oncology, Tata Memorial Hospital, Mumbai, India, between December 2009 and July 2012, were studied. Among these 475 patients, 50 were partially treated and were not in haematological remission. Diagnosis of MM was confirmed by bone marrow pathology and immunobiochemical parameters.
The study protocol was approved by the institutional ethics committee and a waiver to consent was granted.
Interphase FISH: Mononuclear cells from bone marrow aspirate were enriched by Ficoll Hypaque gradient centrifugation. Plasma cells were purified using CD138-coated magnetic beads according to the manufacturer's instructions (Miltenyi Biotec, Paris, France). Enriched plasma cells were identified by fluorescein isothiocyanate-conjugated anti-human kappa lambda light chain staining, and the purity was 95 per cent (range 70-99%).
Interphase FISH was performed on plasma cells using locus-specific probes, LSI 13(D13S319), LSI 13q34 (control), LSI 17(p13.1)(TP53)/CEN 17(D17Z1), LSI break apart dual colour 5’ 3’IgH, dual fusion translocation probes CCND1XT/IgH, FGFR3/IgH, MAF/IgH (Vysis Abbott Molecular, Delkenheim, Germany) and MYC/IgH (Cancer Genetics, Milan, Italy). Hyperdiploidy was analyzed using a set of probes specific for CEP3, EGR1 (5q31)/D5S235(5p12.2), CEP7, CEP11 (Abbott Molecular) and CEP9/15 (Kreatech Diagnostics, The Netherlands). Hyperdiploid MM was defined as the presence of trisomy of ≥2 chromosomes9. FISH procedure was followed as per the manufacturer's protocol. A total 200 interphase plasma cells nuclei were evaluated by two observers. The cut-off threshold for Δ13 [del(13q)/-13], del(17) (p13.1) and t(14q32) using IgH break apart probe was 10 per cent and for dual fusion translocation probes (CCND1XT/IgH, FGFR3/IgH, MAF/IgH, MYC/IgH) and trisomy was five per cent.
Interphase FISH for 1q gain/1q amplification [amp(1q)] was performed on a separate group of 250 newly diagnosed patients (male: 166, female: 84, age 22-86 yr) between October 2012 and December 2013. Locus-specific DNA probes LSI 1q21/1p36 (Kreatech Diagnostics) and LSI 1q25/1p36 (Abbott Molecular) were used for amp(1q) study. The cut-off threshold for amp(1q) was 10 per cent. In this group, amp(1q) was analyzed along with other recurrent cytogenetic markers such as Δ13, del(17) (p13.1), IgH translocations and hyperdiploidy.
Statistical analysis: Of the 475 cases, 392 with proper clinical and laboratory details were enrolled for statistical analysis to investigate correlation with clinical and laboratory features. Of the 392 cases, staging was available in 329 cases (stage I: 89, stage II:122, stage III:118). Association of clinical, haematological and biochemical variables was evaluated by Pearson Chi-square test, Mann-Whitney U-test and independent t test (SPSS, version 18, SPSS Inc., Chicago, USA). Comparison of frequencies among different chromosome abnormality groups was analyzed using proportion test.
Results
Frequency of Δ13, del(17) (p13.1) (TP53 deletion), IgH translocations and hyperdiploidy: Among 475 patients, 312 (66%) had chromosome abnormalities such as trisomies of odd chromosomes 3, 5, 7, 9, 11 and 15, Δ13, del(17) (p13.1) and IgH translocations. The frequencies of chromosome abnormalities were as follows: IgH translocations (128/475, 27%), t(11;14) (26/475, 5%), t(4;14) (47/475, 10%), t(14;16) (16/475, 3%), t(8;14) (5/475, 1%), Δ13 (152/475, 32%), del(17) (p13.1) (41/475, 9%) and hyperdiploidy (173/475, 36%) (Figs 1 and 2). Sole, isolated hyperdiploidy was detected in 20 per cent patients. The median percentage of plasma cells was 80 per cent for all the aberrations. In 60 per cent of cases with del(17p), the plasma cell clone size of TP53 deletion was smaller (20-50%) than IgH clone size (25-99%).

-
(A) CEP 3 probe on interphase cells shows trisomy 3 (3 red signals). (B) CEP 7 probe on interphase cells shows trisomy 7 (3 green signals). (C) LSI 15q22/9q34 probe on interphase cells shows tri-tetrasomy 9 (3-4 red signals) and tetrasomy 15 (4 green signals). (D) LSI 13S319 (13q14.3) and LSI 13q34 (control) on interphase cells show monoallelic deletion of locus 13q14.3 (1 red signal) and two copies of locus 13q34 (2 green signals). (E) LSI 13S19 (13q14.3) and LSI 13q34 (control) on interphase cells show monosomy 13 (1 red signal and 1 green signal). (F) LSI 17p13.1 (TP53)/CEN 17 probe on interphase cells shows monoallelic deletion of TP53 (1 red signal and 2 green signals of CEP 17). (G) LSI 1q21/SRD(1p36) on interphase cells shows amplification of locus 1q21(3 green signals) and 2 copies of locus 1p36 (2 red signals). (H) LSI 1q21/1p36 on interphase cells shows amplification of locus 1q21 (7 green signals) and 2 copies of locus 1p36 (2 red signals). (I) LSI 1q25/1p36 on interphase and metaphase cells shows amplification of locus 1q25 (3 green signals) and two copies of locus 1p36 (2 red signals).
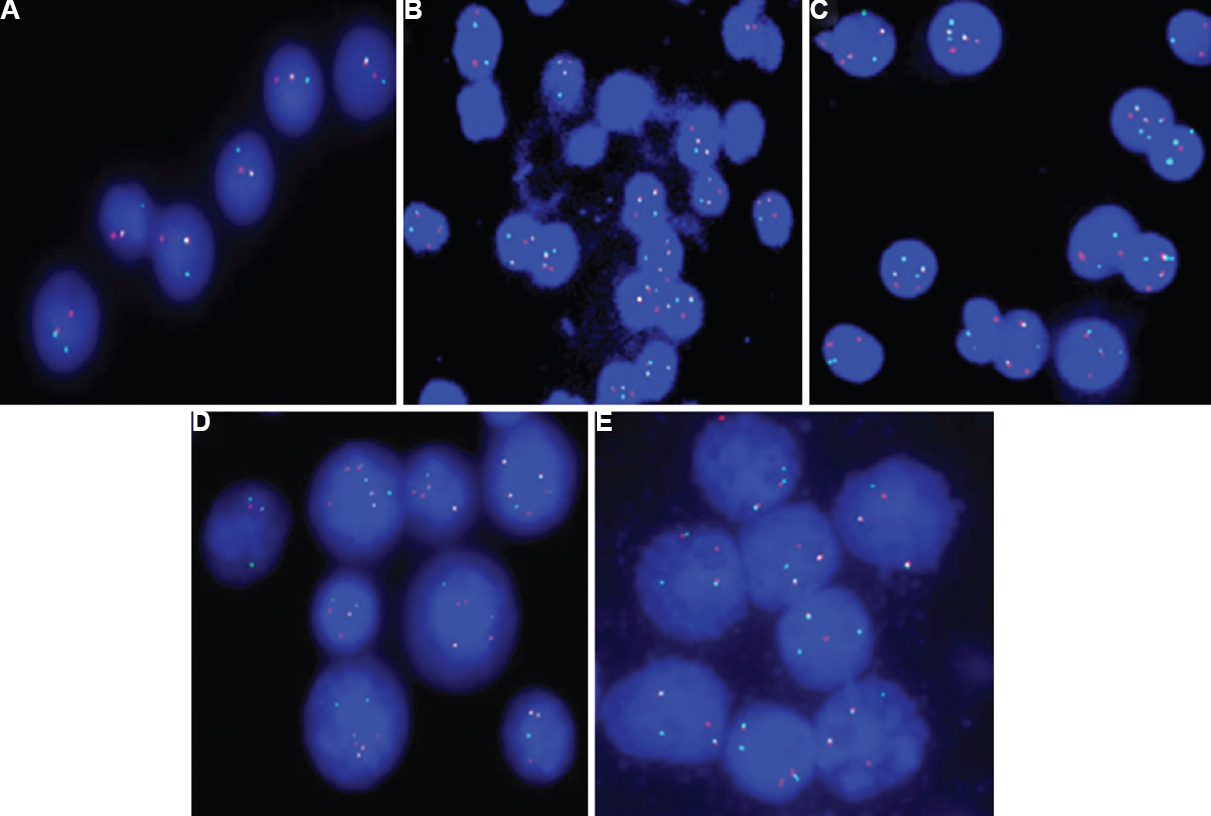
-
(A) Dual colour IgH break apart probe on interphase cells shows normal IgH allele (yellow signal) and residual IgH (1 red signal and 1 green signal). (B) LSI IgH/CCND1 XT dual fusion probe on interphase cells shows dual fusion of IgH-CCND1 (1R1G2Y). (C) LSI IgH/FGFR3 dual fusion probe on interphase cells shows one copy of FGFR3 (1 red signal), two copies of IgH (2 green signals) and 2-3 copies of IgH-FGFR3 (2-3 yellow signals). (D) LSI IgH/MAF dual fusion probe on interphase cells shows 1-2 copies of MAF (1-2 red signals), 1-2 copies of IgH (1-2 green signals) and 1-3 copies of fusion signals of IgH-MAF (1-3 yellow signals). (E) LSI IgH/MYC dual fusion probe on interphase cells shows normal allele of MYC (1 red signal), normal allele of IgH (1 green signal) and dual fusion signal of IgH-MYC (2 yellow signals).
Clustering of Δ13 (P<0.001), del(17) (p13.1) (P<0.001), IgH translocations (P<0.001), t(11;14) (P<0.001), t(4;14) (P<0.001) and t(14;16) (P<0.001) was detected in non-hyperdiploid group (Fig. 2). A strong association was noted between t(4;14) and Δ13/del(17) (p13.1) aberrations in comparison with t(11;14), t(14;16) and other IgH variants (P<0.008).
Clinical, haematological and immunobiochemical significance of cytogenetic aberrations: Of the 475 patients, 392 with proper clinical and laboratory parameters were enrolled for statistical analysis. Patients with Stage I and Stage II were grouped together, and cytogenetic findings of International Staging System (ISS)10 Stages I and II group were compared with Stage III. The occurrence of overall cytogenetic abnormalities as well as individual chromosomal abnormality such as Δ13, del(17) (p13.1), IgH translocations and trisomies of 3, 5, 7, 9, 11, 15 was similar in the group of 392 patients as observed in 475 patients. No difference was observed in the frequencies of various cytogenetic groups of newly diagnosed and partially treated MM cases.
Patients with t(4;14) had low haemoglobin (Hb) (P<0.001), low level of albumin (P<0.001), high ß2-microglobulin (P<0.001), high percentage of plasma cells (P<0.001) and Stage III disease (P<0.001) (Table). High level of ß2-microglobulin (P<0.016) with progressive disease (P<0.007) was observed in patients with del(17) (p13.1). Isolated group of hyperdiploidy was associated with low plasma cell index (P<0.004). Although higher age group was noted in all cytogenetic groups, it was significantly associated with t(14;16) (P<0.025) and hyperdiploid group (P<0.004) (Table).
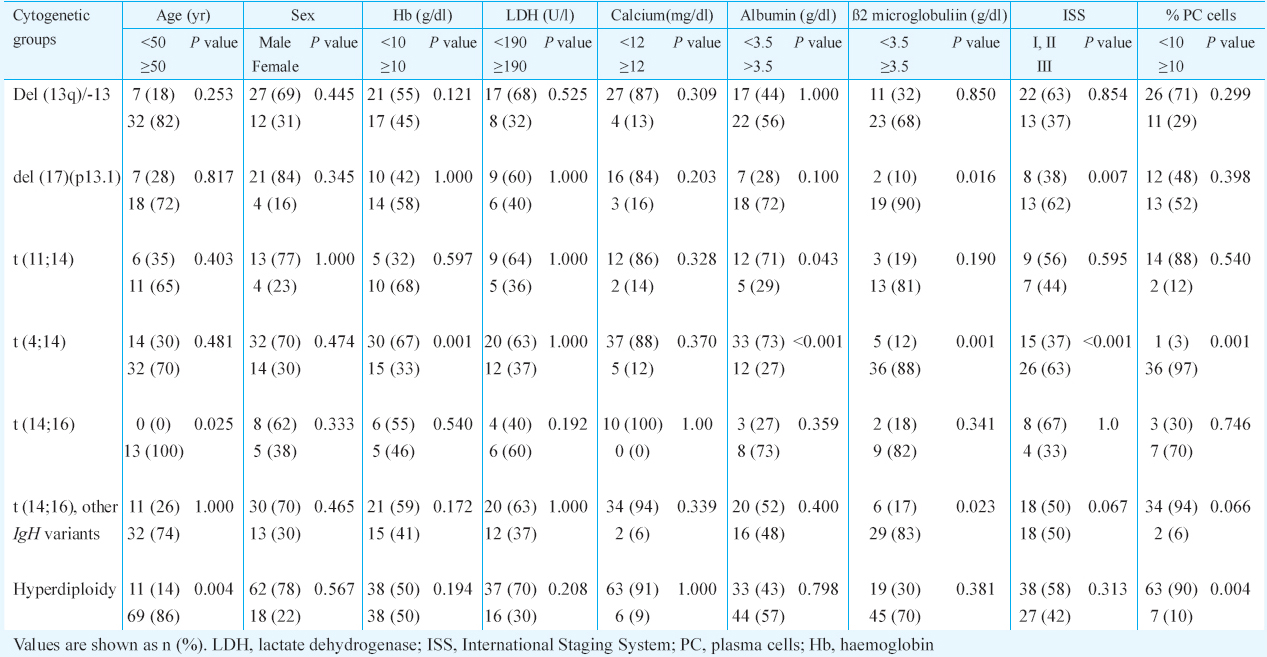
Comparable differences were noted between aberrations positive group versus aberrations negative group. Aberrations positive group had low Hb (P<0.001), low albumin (P<0.001), high ß2-microblobulin (P<0.001), high index of plasma cells (P<0.001) and ISS Stage III (P<0.001).
Chromosome 1 aberrations: In a group of 250 patients, 71 per cent (78/250) had chromosome aberrations and amp(1q) was seen in 83 of 250 (33%) patients. Although 1q21 probe was applied in all patients, 130 patients were also studied both by LSI 1q21 and LSI 1q25 probes, and the amplification at both 1q21 and 1q25 loci was detected with almost same clone size in positive cases (Fig. 1). An isolated amp(1q) was seen in only four per cent cases. On the contrary, amp(1q) was always noted in combination with other recurrent chromosome abnormalities. Of the 250 patients, 184 with proper clinical and laboratory details were enrolled for statistical analysis. A significant association of amp(1q) was seen in patients with t(4;14) (P < 0.03) and t(14;16) (P < 0.05). Clinico-pathological evaluation revealed association of t(4;14) with Δ13 and/or del(17) (p13.1) plus amp(1q) with high ß2-microglobulin (P<0.04), whereas sole Δ13 accompanied by amp(1q) had high ß2-microglobulin (P<0.05) and increased plasma cell index (P<0.002).
Discussion
In the present group of 475 MM patients, interphase FISH could detect recurrent genomic abnormalities in 66 per cent patients, the frequency of which was comparable (50-90%) to that reported in literature14511121314. Various cytogenetic subgroups such as hyperdiploidy, Δ13, del(17) (p13.1) IgH translocation subtypes t(11;14), t(4;14), t(14;16), t(8;14) and amp(1q) were identified. Overall frequencies of del(17) (p13.1) (9%), t(4;14) (10%), t(14;16) (3%), t(8;14) (1%) and hyperdiploidy (36%) were comparable to the reported studies, whereas frequencies of Δ13 (32%) and IgH translocations (27%) were low in comparison with reported studies124511121314. The occurrence of t(11;14) (5%) was significantly lower than t(4;14) (10%) in contrast to reported frequency (15-20% and 10-15%, respectively)11516. The low frequency of t(11;14) lowered the frequency of IgH translocations. This could be due to geographic heterogeneity with ethnical differences. Greenberg et al17 have reported overall low frequency of t(11;14) being 6.5 per cent in African-American Blacks compared with Whites (17.6 %).
Variant IgH translocations other than t(4;14) and t(14;16) are rare subtypes with a frequency of 2-5 per cent, and the targeted IgH loci are C-MYC at q24, CCND3 (Cyclin D3) at 6p21, IRF4 (Interferon regulatory factor 4) at 6p25, MAFB (Musculoaponeurotic fibrosarcoma oncogene homolog B) at 20q111415181920. The prognostic significance of these IgH translocations is not clear due to lack of valid substantial data influencing clinical features and therapeutic response.
As evident from literature145911152122, IgH translocations, Δ13, del(17) (p13.1) were significantly associated with non-hyperdiploid group in our patients. Only a few patients with Δ13 showed heterozygous as well as homozygous deletion which indicated tumour heterogeneity. Association of Δ13/del(17) (p13.1) with t(4;14) in 475 patients and association of amp(1q) with t(4;14) and t(14;16) in the group of 250 patients indicated that del(17) (p13.1) and amp(1q) were progressive events in t(4;14) positive MM.
In a group of 392 patients with clinico-laboratory parameters in agreement with literature reports, hyperdiploid group was low-risk group123411212223. The various recurrent aberrations such as Δ13, TP53 deletion, IgH translocations and hyperdiploidy tended to occur in the old age group (>60 yr). Association of old age MM with higher rate of IgH translocations has been already reported by Butler et al12. Various studies have revealed conflicting findings regarding the relationship of recurrent cytogenetic subtypes and their association with age122425. Evaluation of clinico-pathological impact of various recurrent cytogenetic groups revealed association of t(4;14) and del(17) (p13.1) with high-risk features such as ß2-microglobulin, per cent plasma cells and advanced disease which confirmed that t(4;14) and del(17) (p13.1) were high-risk prognostic groups of MM. Translocation 4;14 and del(17) (p13.1) confer an adverse prognosis with shorter survival1234581819202627. Sole, hyperdiploid MM was low-risk group. On the other hand, clustering of aggressive subtypes t(4;14) and del(17) (p13.1) in non-hyperdiploidy confirmed prognostic impact of ploidy and supported that hyperdiploidy and non-hyperdiploidy were the two major groups of MM148911.
Although 1q21 locus is a hot spot for amp(1q), yet several studies have also reported the involvement of 1q12-1q23. The amp(1q) seen in 250 patients revealed amplification at region 1q21 and 1q25. This indicates the importance of genes underlying the pathogenesis which are scattered in 1q12-1q25 region and not restricted to only 1q2152829303132. The copy number of 1q21 and median percentage of cells with amp(1q21) were found to have increased (more than three copies) in patients with advanced disease. Similar observation has been noted by Hanamura et al28. They found that relapsed patients with four copies of 1q21 at relapse had inferior post-relapse survival compared to those with three copies of 1q21.
Since amp(1q) always occurred in combination with other recurrent aberrations, it was difficult to assess its clinico-pathological impact as a sole or isolated abnormality; however, amp(1q) accompanied by t(4;14) and Δ13 and their association with high-risk features such as high ß2-microglobulin, increased plasma cell index, and advanced disease indicated its progressive role with genomic instability in myeloma pathogenesis. Wu et al29 have also reported association of chromosome 1q aberration with Δ13 with adverse prognosis in patients on high-dose chemotherapy in MM. Though Δ13 was found to be low risk, its adverse prognostic implication could be due to its association with high-risk group such as t(4;14) and amp(1q).
In conclusion, the comprehensive cytogenetic data on MM patients by interphase FISH enabled identification of various prognostic subsets with risk stratification as a part of standard care. Low frequency of chromosome 13 aberrations and t(11;14) was an interesting observation, probably indicating ethnic diversity. Sole hyperdiploidy was a low-risk group, and clustering of aggressive subtypes t(4;14) and del(17) (p13.1) in non-hyperdiploid group confirmed that hyperdiploidy and non-hyperdiploidy were two major prognostic groups of MM. Amp(1q) accompanied by t(4;14) and Δ13 and their association with high-risk clinical and laboratory features focused on progressive role of amp(1q) in MM. Amp(1q) was clustered at 1q21 and also at 1q25 locus. Both interphase FISH with comprehensive profiling of various cytogenetic markers and selective molecular profiling may further improve classification and contribute in risk stratification of disease which should be adapted in standard routine care of MM patients in clinical practice.
Conflicts of Interest: None.
References
- Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546-58.
- [Google Scholar]
- International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210-21.
- [Google Scholar]
- Revised international staging system for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015;33:2863-9.
- [Google Scholar]
- Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood. 2007;109:3489-95.
- [Google Scholar]
- The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011;204:3-12.
- [Google Scholar]
- Cytogenetic abnormalities correlate with the plasma cell labeling index and extent of bone marrow involvement in myeloma. Cancer Genet Cytogenet. 1999;113:73-7.
- [Google Scholar]
- Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012;97:1272-7.
- [Google Scholar]
- Molecular heterogeneity of multiple myeloma: pathogenesis, prognosis, and therapeutic implications. J Clin Oncol. 2011;29:1893-7.
- [Google Scholar]
- Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19:275-8.
- [Google Scholar]
- Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585-90.
- [Google Scholar]
- Association of age with fluorescence in situ hybridization abnormalities in multiple myeloma reveals higher rate of IGH translocations among older patients. Leuk Lymphoma. 2012;53:2444-8.
- [Google Scholar]
- Cytogenetic profiles in multiple myeloma and monoclonal gammopathy of undetermined significance: a study in highly purified aberrant plasma cells. Haematologica. 2013;98:279-87.
- [Google Scholar]
- Impact of cytogenetics on clinical outcome in multiple myeloma: a risk stratification model. J Clin Oncol. 2011;29:Suppl. Abstract e18576
- [Google Scholar]
- t(11;14) and t(4;14) translocations correlated with mature lymphoplasmacytoid and immature morphology, respectively, in multiple myeloma. Leukemia. 2003;17:2032-5.
- [Google Scholar]
- Racial differences in primary cytogenetic abnormalities in multiple myeloma: a multi-center study. Blood Cancer J. 2015;5:e279.
- [Google Scholar]
- Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid patients. Genes Chromosomes Cancer. 2003;38:234-9.
- [Google Scholar]
- Molecular classification and risk stratification of myeloma. Hematol Oncol. 2013;31(Suppl 1):38-41.
- [Google Scholar]
- Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood. 2001;98:217-23.
- [Google Scholar]
- Prognostic factors for hyperdiploid-myeloma: effects of chromosome 13 deletions and IgH translocations. Leukemia. 2006;20:807-13.
- [Google Scholar]
- Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012;119:2100-5.
- [Google Scholar]
- Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229-38.
- [Google Scholar]
- Chromosomal abnormalities of young multiple myeloma patients (<45 yr) are not different from those of other age groups and are independent of stage according to the international staging system. Eur J Haematol. 2007;78:227-34.
- [Google Scholar]
- Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia. 2005;19:1634-42.
- [Google Scholar]
- The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808-22.
- [Google Scholar]
- Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837-40.
- [Google Scholar]
- Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724-32.
- [Google Scholar]
- Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007;136:615-23.
- [Google Scholar]
- Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology. 2005;10(Suppl 1):117-26.
- [Google Scholar]
- Overexpression of PDZK1 within the 1q12-q22 amplicon is likely to be associated with drug-resistance phenotype in multiple myeloma. Am J Pathol. 2004;165:71-81.
- [Google Scholar]
- 1q21 amplification with additional genetic abnormalities but not isolated 1q21 gain is a negative prognostic factor in newly diagnosed patients with multiple myeloma treated with thalidomide-based regimens. Leuk Lymphoma. 2012;53:2500-3.
- [Google Scholar]






