Translate this page into:
Evaluation of colorimetric nitrate reductase assay for rapid detection of methicillin resistance in clinical isolates of Staphylococcus aureus
Reprint requests: Dr Sujatha Sistla, Professor, Department of Microbiology, Jawaharlal Institute of Postgradute Medical Education & Research, Puducherry 605 006, India e-mail: sujathasistla@gmail.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Methicillin resistant Staphylococcus aureus (MRSA) remains a major cause of health care-associated infections. Rapid detection of MRSA facilitates the early initiation of appropriate treatment and infection control. Hence, the present study was undertaken to standardize and evaluate the performance of rapid colorimetric nitrate reductase assay (NRA) for determining methicillin resistance in S.aureus.
Methods:
A total of 160 clinical isolates of S. aureus, (80 each of methicillin susceptible and methicillin resistant) were included in the study. Minimum inhibitory concentration (MIC) was determined by NRA and reference broth micro dilution (BMD) methods. Results of NRA were compared with BMD and analyzed.
Results:
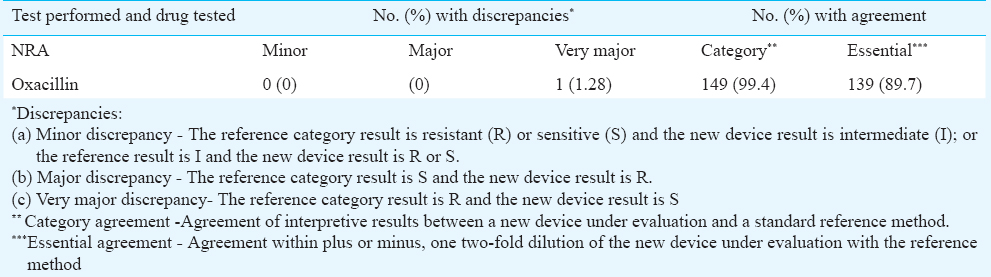
For MRSA, the MIC values ranged from 4 to ≥ 16 μg/ml and for MSSA, ≤ 0.5 to 2 μg/ml. Category and essential agreement for NRA as compared with BMD were found to be 99.4 and 89.7 per cent, respectively. No minor or major discrepancy was observed. A single resistant isolate showed very major discrepancy.
Interpretation & conclusions:
Colorimetric NRA being an inexpensive test requiring no special equipment can be employed as an alternative method for rapid detection of MRSA in resource limited settings.
Keywords
Broth microdilution
MRSA
nitrate reductase assay
rapid detection
Methicillin resistant Staphylococcus aureus (MRSA) is known to be a major cause of healthcare-associated infections and also community-acquired infections despite the availability of effective preventive and therapeutic strategies. Methicillin resistance in S. aureus is mediated by the production of PBP2a protein, an altered penicillin-binding protein with lower affinity for beta-lactam antibiotics than PBP2, which is encoded by the mecA gene1. Though several methods for MRSA detection have been developed, these are either slow or not sufficiently sensitive or specific2. Cefoxitin disc diffusion and oxacillin screen agar are the currently recommended phenotypic methods by the Clinical and Laboratory Standards Institute (CLSI), where the results are available only after 24 h3. Although this may not have serious impact on patient management in most cases, yet such a delay may worsen the outcome in life-threatening infections like sepsis, endocarditis, meningitis, osteomyelitis, and catheter-related infections. In such situations, rapid identification of MRSA is of paramount importance. The two rapid tests which are widely employed in developed countries, are PCR for mecA gene4 and latex agglutination test for PBP2a4, but these are not affordable in most resource limited laboratories.
Determining the minimum inhibitory concentration (MIC) values by the reference broth micro dilution (BMD) method is inexpensive but laborious and time consuming. The colorimetric nitrate reductase assay (NRA) combines the rapidity of PCR and low cost of BMD5. NRA is a biochemical test based on the reduction of nitrate to nitrite by viable bacteria and a colour change produced by the addition of a reagent. NRA has been extensively evaluated in the past for the drug susceptibility in Mycobacterium tuberculosis6. In addition to NRA, other colorimetric tests like XTT 2, 3-bis-(2-methoxy-4-nitro-5-sulphophenyl)-2H-tetrazolium-J-carboxanilide] assay, which is based on the reduction of a tetrazolium salt has been developed and evaluated for rapid antimicrobial susceptibility testing in Pseudomonas aeruginosa7. The present study was carried out to standardize and evaluate the performance of NRA in comparison with BMD and PCR to detect the methicillin resistance among clinical isolates of S. aureus.
Material & Methods
The study was carried out in the department of Microbiology, Jawaharlal Insititute of Postgraduate Medical Education and Research (JIPMER), Puducherry, India, for a period of two months (March and April, 2013) after obtaining approval from the Institute Research and Human Ethics committees.
Bacterial isolates: For standardization of NRA, American Tissue Culture Collection (ATCC) Staphylococcus aureus 29213 (methicillin susceptible) and ATCC S. aureus 43300 (methicillin resistant) were included. A total of 160 clinical isolates of S. aureus [80 each of methicillin sensitive (MS) SA and MRSA] were included for evaluation. These isolates were obtained from blood (total=29; MRSA- 12; MSSA -17), pus and exudates (total=108; MRSA -61; MSSA-47), CSF and other sterile body fluids (total =12; MRSA-3; MSSA-9), tracheal aspirate (total =11; MRSA-4; MSSA-7).
Detection and confirmation of MRSA: Methicillin resistance among clinical isolates of S. aureus was detected using oxacillin screen agar (6 μg/ml) and cefoxitin (30 μg) disc diffusion according to CLSI recommendations3. MIC values of oxacillin were also determined by the broth microdilution method according to CLSI recommendations3. Oxacillin was tested in concentrations of 8 to 0.25 μg/ml. Breakpoints of MICs for susceptibility and resistance were defined by CLSI, as ≤2 μg/ml and ≥4 μg/ml, respectively. To confirm methicillin resistance in MRSA isolates, PCR for mecA gene (310 bp) was carried out using primers8, forward 5’-TAGAAATGACTGAACGTCCGATAA-3’ and reverse 5’-CCAATTCCACATTGTTTCGGTCTAA-3’ and amplification conditions as follows: initial denaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 45 sec, annealing at 50°C for 45 sec, extension at 72°C for one min and final extension step at 72°C for three minutes8.
Performance of NRA: The assay was performed with some modifications of the CLSI recommended broth microdilution method5 using U- bottomed 96-well microtitre plates. The bacterial suspension was prepared from overnight culture in cation-adjusted Mueller-Hinton (M-H) broth with 1,000 μg/ml potassium nitrate and 4 per cent NaCl. After adjusting to 0.5 McFarland turbidity standard, 50 μl of bacterial suspension was added to each well containing 50 μl of antibiotic solution, including control (antibiotic-free) well such that the final oxacillin concentration ranged from 8 to 0.25 μg/ml. Microtitre plates were covered and incubated at 35°C. Fifty micro litres of freshly prepared Griess reagent [2 units of 0.1% N-(1-naphthyl) ethylene diamine and 2 units of 0.2 per cent sulphanilamide in distilled water and 1 unit of 50 per cent hydrochloric acid, Hi-Media, Mumbai] was added into control wells at the fifth hour of incubation. After the addition of Griess reagent, if a violet- purple colour change was seen in the control well, it was considered that sufficient bacterial growth has occurred. The reagent was then added to the other wells and the colour change, if any was noted. The MIC was defined as the lowest drug concentration without colour change. In each batch of test, growth control well and sterility control well were included to check the microbial growth and the sterility of microtitre plate, respectively. In addition, ATCC S. aureus 29213 (methicillin susceptible) and ATCC S. aureus 43300 (methicillin resistant) were included for quality control5.
Test for reproducibility: The test was carried out in triplicate, with the ATCC strains and 60 test isolates of S. aureus (30 of each MSSA and MRSA), on three different days. To remove observer bias, the investigator performing and interpreting the NRA results was blinded to the identity of the methicillin susceptibility status of the test isolates.
Results were analyzed according to FDA (Food and Drug Administration) criteria9. Minor, major, and very major discrepancy rates and category and essential agreement rates were determined. In addition, sensitivity, specificity, positive and negative predictive values of NRA for the detection of MRSA were calculated.
Results
All 80 MSSA isolates were susceptible to cefoxitin by disc diffusion and oxacillin by oxacillin screen agar and all MRSA isolates were resistant to both. There was 100 per cent concordance in the results of the two phenotypic tests (oxacillin screen agar and cefoxitin disc diffusion) for the identification of MSSA and MRSA. All 80 MRSA isolates were positive for mecA gene by PCR.
Results of NRA when tested with ATCC S. aureus 29213 (methicillin susceptible) and ATCC S. aureus 43300 (methicillin resistant) were comparable with those of reference BMD method. Although one of the triplicates of the susceptible isolate showed one well difference, final interpretation remained unchanged.
On checking for reproducibility with the 60 test isolates, identical MIC values were obtained in 12 isolates by NRA and in 19 by reference BMD method. The remaining isolates showed the same MIC reading on at least two occasions.
Table I shows the distribution of MICs determined by colorimetric NRA and BMD method for the test isolates. For MRSA, the MIC values ranged from 4 to ≥16 μg/ml and for MSSA, ≤ 0.5 to 2 μg/ml. Of the 160 test isolates, five (2 – MRSA, 3 – MSSA) did not show colour change at the end of the fifth hour and hence could not be included for evaluation.

Category and essential agreement for NRA were found to be 99.4 and 89.7 per cent, respectively, in comparison with the reference method. No minor and major discrepancy was observed. However, one of the 78 resistant isolates (1.28%) showed very major discrepancy as depicted in Table II.
Sensitivity, specificity, positive and negative predictive values of NRA for the detection of MRSA were found to be 98.7, 100, 100 and 98.7 per cent respectively.
Discussion
Colorimetric NRA based on the ability of bacteria to reduce nitrates to nitrites is a rapid, inexpensive method which is easy to perform. Coban et al2, used this method for the detection of methicillin and vancomycin resistance among isolates of S. aureus5. In their study, there was an absolute agreement of 100 per cent and essential agreement of 91.6 per cent between NRA and BMD, although no minor, major or very major discrepancies were observed for oxacillin. Results of our study showed a broad agreement with those of Coban et al5, except for the slightly lower essential agreement and occurrence of very major discrepancy with a single isolate. According to the guidelines issued by FDA9, essential agreement should be 95.33 per cent, when the number of isolates tested is 150. In our study, the essential agreement was 89.67 per cent, which is less than the recommended level.
Any test which is used to determine antibiotic susceptibility of organisms cannot afford to misidentify a resistant isolate as a sensitive one. This constitutes a very major discrepancy according to FDA criteria and has serious clinical implications. In the present study there was such an error with a single isolate. One possible reason for this error could be the very small difference in the MIC values of oxacillin for sensitive vs. resistant isolates (2 and 4 μg/ml, respectively), which completely changes the interpretation even with a single well difference in the test. Whether this would severely limit the usefulness of NRA remains to be seen.
Acuner and Eroglu10 commented that the study results reported by Coban et al5, were unacceptable as the reproducibility of NRA was not established. To address this lacuna, we carried out the test in triplicate for 30 isolates, where 12 and 19 isolates showed identical MIC value by NRA and BMD, respectively. Although the reproducibility of NRA was slightly lower than the reference method, it should not completely negate the value of this assay, as the reproducibility of even the reference method was not 100 per cent.
The only problem faced while carrying out this test was that not all the isolates showed colour change in antibiotic-free well, at the end of the fifth hour. In routine practice, there may be some isolates which are slow-growers, for which this test is not applicable. In our study, we encountered five such isolates which could not be included for evaluation.
Other colorimetric tests like resazurin microplate assay (REMA) and alamar blue are also employed for the rapid detection of antimicrobial resistance1112. Advantage of NRA when compared with REMA is that the results are obtained in five hours with the former, whereas in the latter, results are obtained after six hours512. To make NRA more user-friendly and to put it into routine use, the breakpoint susceptibility testing alone can be carried out instead of determining the entire range of MIC values, which was illustrated in a recent report by Coban et al13.
Rapid antimicrobial susceptibility has also been determined by automated systems such as BD Phoenix and Vitek 2 in clinical microbiology laboratories14. Unfortunately, like PCR for mecA gene and latex agglutination test (LAT) for PBP2a, these are also expensive and can be used only in well-equipped laboratories. In such a scenario, NRA can be an alternative method, as it does not require any expensive reagents or equipment. NRA can also be employed for any other bacteria- antibiotic combination, provided the organisms are capable of reducing nitrates to nitrites.
To conclude, colorimetric NRA is easy to perform and saves time in the detection of MRSA, as the identification of S. aureus and the status of their methicillin susceptibility can be determined on the same day, which helps the clinician to start appropriate antibiotic therapy. Testing a large number of clinical isolates of S. aureus and MRSA for oxacillin resistance in different laboratories will help to further validate this assay.
Conflicts of Interest: None.
References
- Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265-73.
- [Google Scholar]
- Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. J Antimicrob Chemother. 2005;55:379-82.
- [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 23rd Informational Supplement. M100-S22. Wayne, PA: CLSI; 2013.
- Diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus in Latin America. Braz J Infect Dis. 2010;14(Suppl 2):S97-106.
- [Google Scholar]
- Two new colorimetric methods for early detection of vancomycin and oxacillin resistance in Staphylococcus aureus. J Clin Microbiol. 2006;44:580-2.
- [Google Scholar]
- Rapid and inexpensive drug susceptibility testing of Mycobacterium tuberculosis with a nitrate reductase assay. J Clin Microbiol. 2002;40:553-5.
- [Google Scholar]
- Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48:1879-81.
- [Google Scholar]
- Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27:27-9.
- [Google Scholar]
- 2009. Food and Drug Administration. Guidance for industry and FDA. Class II special controls guidance e document: antimicrobial susceptibility test (AST) systems. Center for Devices and Radiological Health, Food and Drug Administration, U.S. Department of Health and Human Services. Available from: http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm388961.pdf
- Unacceptable performance and the lack of reproducibility results in the report of colorimetric methods for early detection of vancomycin and oxacillin resistance in Staphylococcus aureus. J Clin Microbiol. 2006;44:2318-9.
- [Google Scholar]
- Evaluation of Alamar colorimetric broth microdilution susceptibility testing method for staphylococci and enterococci. J Clin Microbiol. 1996;34:2654-9.
- [Google Scholar]
- Rapid determination of methicillin resistance among Staphylococcus aureus clinical isolates by colorimetric methods. J Clin Microbiol. 2012;50:2191-3.
- [Google Scholar]
- Nitrate reductase assay for the rapid detection of Staphylococcus aureus methicillin resistance: A breakpoint susceptibility testing method. Mikrobiyol Bul. 2014;48:40-7.
- [Google Scholar]
- Use of the BD PHOENIX Automated Microbiology System for direct identification and susceptibility testing of gram-negative rods from positive blood cultures in a three-phase trial. J Clin Microbiol. 2004;42:1466-70.
- [Google Scholar]






