Translate this page into:
Dual transcripts of BCR-ABL & different polymorphisms in chronic myeloid leukaemia patients
Reprint requests: Dr Soumen Chakraborty, Institute of Life Sciences, Nalco Square, Bhubaneswar 751 023, Odisha, India e-mail: soumen_ils@yahoo.co.in, soumen@ils.res.in
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Chronic myeloid leukaemia is (CML) characterized by the presence of a hallmark chromosomal translocation, the Philadelphia chromosome. Although there are many reports available regarding the different variants of BCR-ABL in CML, we studied the co-expression of e13a2 and e14a2 transcripts and a few polymorphisms in CML patients.
Methods:
Molecular genetics approach was adapted to screen for polymorphisms, mutation and translocation in BCR, ABL kinase domain and BCR-ABL breakpoint region in 73 CML patients.
Results:
All eight patients with dual transcripts were found to harbour an exonic polymorphism (c.2700 T>C) and an intronic polymorphism (g.109366A>G) that were earlier reported to be associated with co-expression of both the transcripts. We also observed c.763G>A mutation in ABL kinase domain and two polymorphisms, c.2387 A>G and c.2736A>G in the BCR gene.
Interpretation & conclusions:
Though our data support the previous findings that co-expression of BCR-ABL transcripts is due to the occurrence of exonic and intronic polymorphisms in the BCR gene, it also shows that the intronic polymorphism can arise without the linked exonic polymorphism. The occurrence of ABL kinase domain mutation is less frequent in Indian population.
Keywords
ABL kinase domain
bipolar disorder
chronic myeloid leukaemia
dual transcript
Philadelphia chromosome
polymorphism
Chronic myeloid leukaemia (CML), a clonal disorder of the haematopoietic stem cells is characterized by the presence of Philadelphia (Ph) chromosome. The Ph chromosome is generated as a result of reciprocal translocation between the long arms of chromosome 9 and 22 wherein, a part of Abelson (c-ABL) gene from chromosome 9 fuses with the Breakpoint Cluster Region gene (BCR) of chromosome 22, resulting in the formation of BCR-ABL oncogene. About 95 per cent of CML patients have Ph chromosome and it has been reported to be present in 25-30 per cent of acute lymphoid leukaemia (ALL) and occasionally in acute myeloid leukaemia (AML) patients1. The BCR-ABL oncogene is translated into BCR-ABL oncoprotein that shows constitutive tyrosine kinase activity1. Depending on the breakpoint in the BCR gene, different transcripts of BCR-ABL oncogene are reported. It was observed that most of the breakpoints involve exon 13 or exon 14 of the BCR major (M-BCR) region which fuses with the exon 2 of ABL gene resulting in e13a2 or e14a2 mRNA that gets translated into p210 kDa BCR-ABL oncoprotein1. There are also reports with breakpoints in exon 1 of BCR minor (m-BCR) region giving rise to e1a2 transcript that gets translated into a p190 kDa oncoprotein. The p190 kDa oncoprotein is mostly related to ALL1. There are a few reports where the breakpoint occurs in the exon 19 of BCR micro (μ-BCR) region giving rise to e19a2 transcript which encodes for a 230 kDa oncoprotein which comprises CML with neutrophilic predominance (CML-N)23. Other transcripts such as b2a3, b3a3, e1a3, e6a2 or e2a2 have also been reported4.
The occurrence of dual transcripts has been linked to two polymorphisms, a thymine (T) to cytosine (C) in exon 13 of BCR gene altering the third nucleotide in the relevant codon (AAT to AAC) resulting in a silent mutation without altering the amino acid (Asp to Asp) and the other one is adenine (A) to guanine (G) polymorphism within intron 13 of the BCR gene5. Branford et al5 have earlier proposed a mechanism for the co-expression of both the transcripts. The intronic polymorphism that occurs at invariant A is a poorly conserved breakpoint that results in reduced efficiency of exon 14 skipping and alternative transcription of BCR and BCR-ABL alleles. This further results in the activation of a cryptic branchpoint that leads to reduced efficiency of BCR intron 13 splicing and BCR-ABL allele and alternatively leading to the use of acceptor site at the end of intron 14, hence resulting in the co-expression of both the transcripts5.
Here we report the co-expression of e13a2 and e14a2 transcripts, a kinase domain mutation and a set of polymorphisms in the BCR gene, which are also known to be associated with other diseases, in CML patients.
Material & Methods
A total of 73 consecutive patients diagnosed with CML36 (70-chronic phase, 1-accelerated phase and 2-blast crisis phase) from January 2013 to March 2015, and five healthy controls (family members of patients who gave consent) were included in the study (38 patients from Sparsh Hospital & Critical Care Unit, Bhubaneswar and 35 from SCB Medical College, Cuttack). Written informed consent was obtained from all patients and the study was approved by the ethics committee of the Institute of Life Sciences, Bhubaneswar, Odisha, India. The inclusion criteria included patients with CML above the age of 18 yr. Pregnant women and terminally ill patients were excluded.
DNA extraction, RNA isolation and cDNA preparation: Peripheral blood samples (0.5-0.8 ml) were collected in an EDTA coated vacutainer tube. Genomic DNA was extracted from the peripheral blood of all control and CML patients using Genelute Blood genomic DNA kit (Sigma-Aldrich, USA) following the manufacturer's protocol. RNA was isolated from the peripheral blood of the control and patients using Ambion's mirVana miRNA isolation kit (Life technologies, USA) following the manufacture's instruction. Complementary DNA (cDNA) was prepared from 50-200 ng of RNA using First strand cDNA synthesis kit (Thermo Scientific, USA). PCR primers were designed to amplify the breakpoint region (BCR-ABL) [breakpoint region forward (F) and reverse (R)], e13a2 and e14a2 transcript specific forward primers (BCR exon 13 F and BCR exon 14 F) and a common ABL R primer, respectively to confirm the presence of dual transcript, ABL gene (ABL control F and R), BCR gene (BCR exon 10 F and 13 to check for polymorphisms in BCR region and BCR intron 13 F and BCR exon 14 R to check for the presence of intronic polymorphism) and the ABL kinase domain (First PCR-breakpoint region F and ABL exon 10 R followed by a second PCR using ABL kinase domain F and R primers). The list of primers used in the study is shown in Table I 57891011. Polymerase chain reaction (PCR) was performed using the Eppendorf Mastercycler gradient (Eppendorf Scientific, Germany). For amplification of the breakpoint region (BCR-ABL), BCR gene, ABL gene and ABL kinase domain from the cDNA, the PCR mixture (15 μl) contained 100 mM Tris HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.1 per cent triton X-100, 10 mM dNTPs (deoxynucleotides), 10 pmol of the forward and reverse primers, 2 units of DyNAzyme II DNA polymerase (Thermo Scientific, USA). The products were resolved in agarose gel and the desired amplicons were purified using GenElute gel extraction kit (Sigma-Aldrich, USA) and sequenced on Applied Biosystems Genetic analyzer 3500 (Life Technologies, USA).
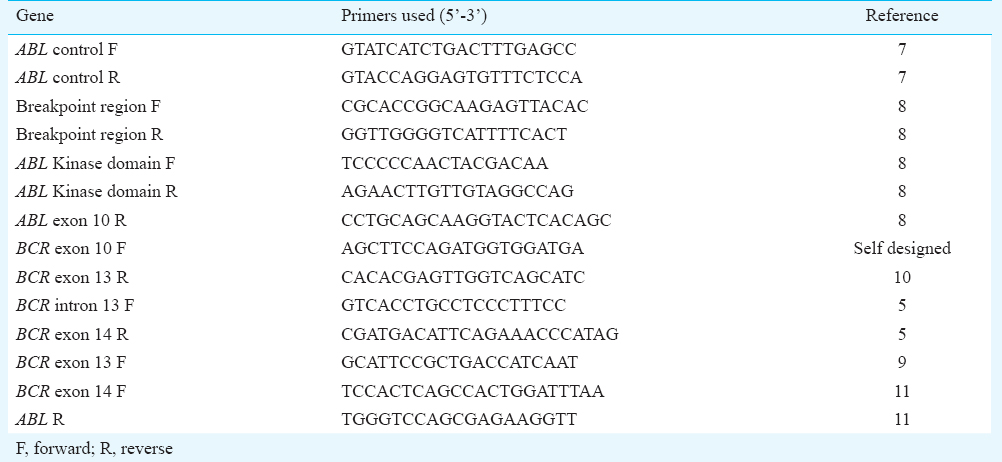
Mutation detection: Sequence data were analyzed using the Chromas Lite ν 2.01 software (http://www.technelysium.com.au/chromas_lite.html) and blast searched using NCBI nucleotide Blast, aligned with reference Genbank cDNA sequence (BCR - NM_004327 and ABL - NM_005157) using EMBL-EBI clustal Omega multiple sequence alignment tool (http://www.ebi.ac.uk/Tools/msa/clustalo/). The disease causing potential of the altered sequences was evaluated using Mutation Taster software tool (http://www.mutationtaster.org). The mutations were checked with already available mutations and polymorphisms in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/).
Results & Discussion
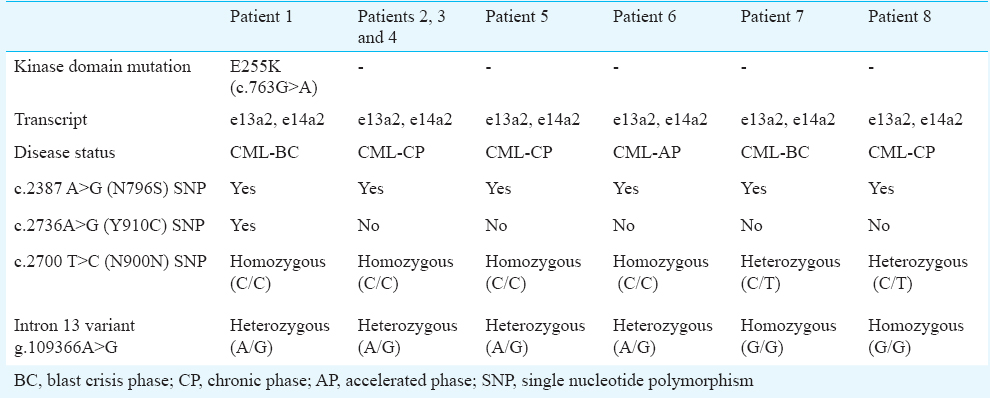
Of the 73 CML patients evaluated, 32 (44 %) were positive for e13a2 and 33 patients were positive for e14a2 (45%) transcript and eight (11%) expressed both e13a2 and e14a2. Of these eight patients, five were in chronic phase (CP), one in accelerate phase (AP) and two in blast crisis phase (BC) (Fig. 1). We investigated for the occurrence of associated polymorphisms in BCR gene in these patients. The exonic polymorphism (2700 T>C, N900N) was observed in 18 of 32 (56.25%) e13a2 patients (homozygous), 5 of 33 (15.15%) e14a2 patients (homozygous) and all eight (100%) patients with dual transcripts (6/8 homozygous condition and 2/8 heterozygous condition) (Fig. 2A). The intronic polymorphism (g.109366A>G) was also observed in all eight patients with dual transcripts (1/8 homozygous condition and 7/8 heterozygous condition) (Fig. 2B). In addition, a few other polymorphisms were also observed in the BCR gene of the patients with dual transcripts; N796S (rs140504) polymorphism in all and Y910C (rs35537221) polymorphism in one patient (Fig. 2C, 2D). As three patients were in the advanced phase of CML, we also looked for possible kinase domain mutation in these patients. An E255K mutation was observed in one of the blast crisis patients (Fig. 2E). This patient also had the N796S and Y910C polymorphism. The list of polymorphisms and mutations that were observed is summarized in Table II (Fig. 2F).
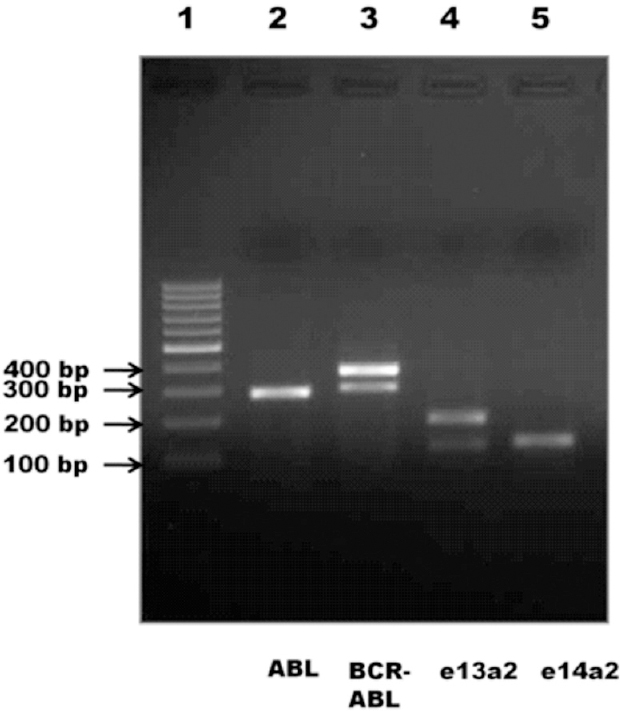
- Agarose gel electrophoresis of dual transcript samples. Lane 1- 100 bp ladder, lane 2- ABL control (288 bp), lane 3- breakpoint region (BCR-ABL) (e13a2-308 bp and e14a2-383 bp), lanes 4 and 5- e13a2 (123 bp and 198 bp) and e14a2 (129 bp) transcript specific PCR, respectively.
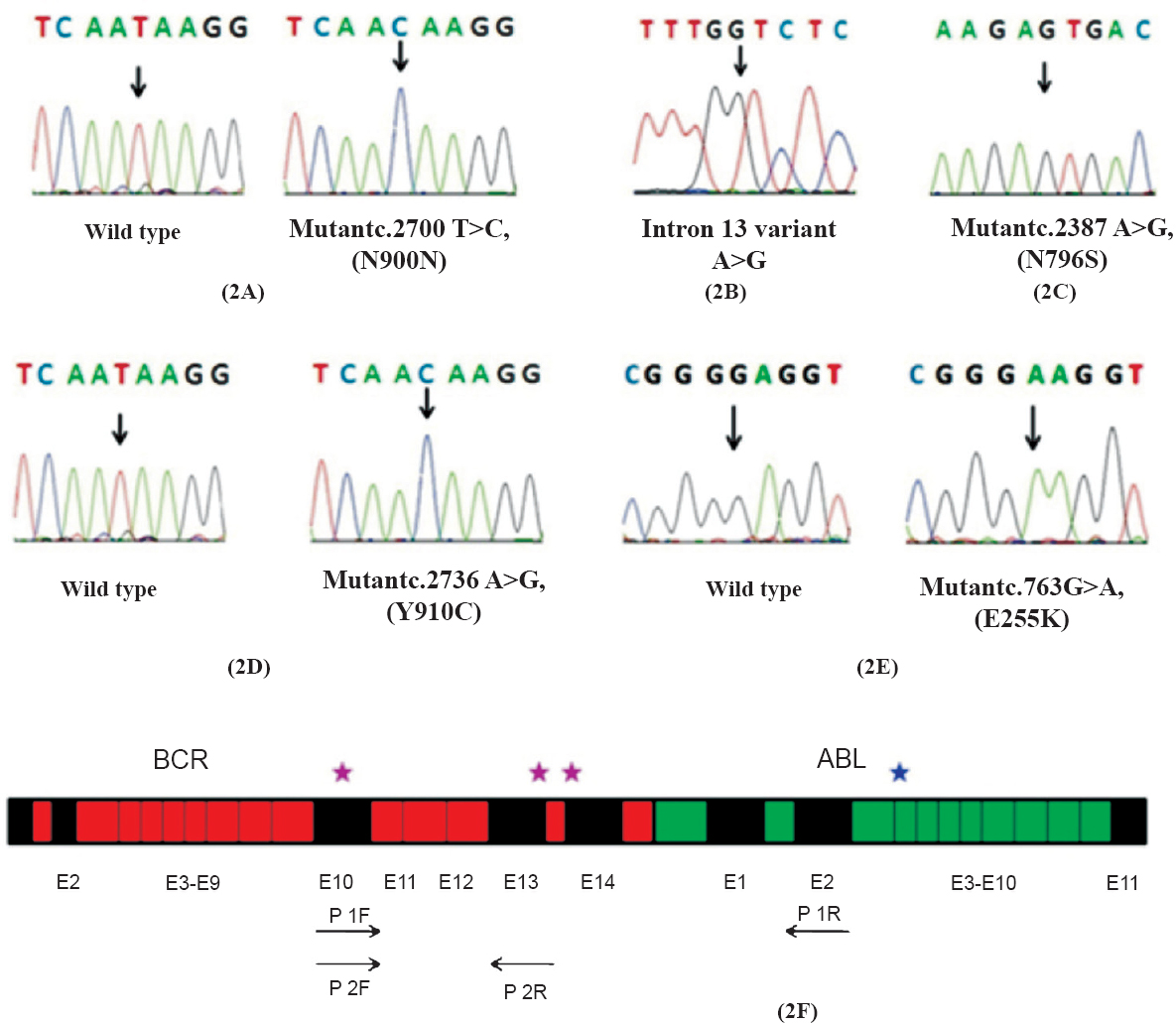
Co-expression of e13a2 and e14a2 transcripts has been reported in certain population groups: 13 per cent of Mexican, 2.5 per cent of Mexican Mestizos, 5.36 per cent of east Indian, 4.6 per cent of Sudanese and 4.38 per cent of north Indian population912131415. The occurrence of dual transcripts is not phase specific as it has been observed in all phases of the disease. Branford et al5, have shown earlier that the presence of BCR's intronic polymorphism is associated with the activation of cryptic splice site that most likely results in reduced efficiency of RNA splicing and the skipping of exon 14 in BCR and BCR-ABL. Mondal et al9 reported six patients with dual transcripts from eastern India. They observed intronic polymorphism in all six individuals at least in heterozygous condition, however, the exonic polymorphism was not observed in all the patients. Although our study included only 73 patients, but the frequency of co-expression of both e13a2 and e14a2 was more when compared to other study groups from India. Our study also shows that the intronic polymorphism arises without the linked exonic polymorphism in the Indian population.
Although the acquisition of BCR-ABL is enough for the initiation of CML, the occurrence of additional secondary mutations is required for the progression of the disease from the initial chronic phase to the fatal blast crisis phase. Several study groups have reported the presence of ABL kinase domain mutations as one of the molecular mechanisms for drug resistance and disease progression16. Kuila et al8, have earlier reported that the occurrence of ABL kinase domain mutation in Indian population is less frequent when compared to other populations.
Hashimoto et al17, in a study in 171 Japanese patients reported that N796S polymorphism was strongly associated with bipolar II disorder. Another study reported that bipolar disorder was associated with interferon-alpha treatment18. Although interferon-alpha was used in CML therapy, we found this particular polymorphism in control samples also. Thus, this polymorphism may not have any association with interferon alpha treatment or bipolar disorder. We also observed Y910C (rs35537221) polymorphism in one patient. Though this polymorphism was predicted to be disease causing from mutation taster prediction tool, it occurs in 1000 Genome Project (TGP) and thus it is likely to be a polymorphism. No possible disease was found to be associated with this polymorphism but it was reported as a germ line variant by Greenman et al19. Isokpehi et al20 have also reported it as a non-synonymous polymorphism that can be used as a candidate marker for arsenic responsiveness in protein targets.
In conclusion, our study shows the frequency of dual transcripts in 11 per cent of our patients. It also suggests that the intronic polymorphism can arise without the linked exonic polymorphism.
Acknowledgment
The study was supported by the Institutional core grant. The first author (SRN) received fellowship from the Department of Science and Technology (DST), and the second author (NK) received DST inspire ICMR-senior research fellowship from Indian Council of Medical Research, New Delhi, India.
Conflicts of Interest: None.
References
- The diversity of BCR–ABL fusion proteins and their relationship to leukaemia phenotype. Blood. 1996;88:2375-84.
- [Google Scholar]
- Neutrophilic-chronic myeloid leukaemia: a distinct disease with a specific molecular marker (BCR/ABL with C3/A2 junction) Blood. 1996;88:2410-4.
- [Google Scholar]
- The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukaemia: rationale and important changes. Blood. 2009;114:937-51.
- [Google Scholar]
- Dual transcription of b2a2 and b3a2 BCR-ABL transcripts in chronic myeloid leukaemia is confined to patients with a linked polymorphism within the BCR gene. Br J Haematol. 2002;117:875-7.
- [Google Scholar]
- The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292-302.
- [Google Scholar]
- SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343-52.
- [Google Scholar]
- Presence of a new BCR-ABL kinase domain mutation, C330G in an imatinib naive patient with chronic myeloid leukemia: very low prevalence of BCR-ABL kinase domain mutations in patients with chronic myeloid leukaemia from eastern India. Leuk Lymphoma. 2009;50:663-6.
- [Google Scholar]
- Molecular profiling of chronic myeloid leukaemia in eastern India. Am J Hematol. 2006;81:845-9.
- [Google Scholar]
- Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747-52.
- [Google Scholar]
- e19a2 BCL-ABL fusion transcript in typical chronic myeloid leukaemia: a report of two cases. J Clin Pathol. 2006;59:1102-3.
- [Google Scholar]
- Analysis of Bcr-abl type transcript and its relationship with platelet count in Mexican patients with chronic myeloid leukaemia. Gac Med Mex. 2003;139:553-9.
- [Google Scholar]
- Frequencies of the breakpoint cluster region types of the BCR/ABL fusion gene in Mexican Mestizo patients with chronic myelogenous leukaemia. Rev Invest Clin. 2004;56:605-8.
- [Google Scholar]
- Frequencies of BCR-ABL1 fusion transcripts among Sudanese chronic myeloid leukaemia patients. Genet Mol Biol. 2010;33:229-31.
- [Google Scholar]
- Cytogenetic & molecular analyses in adult chronic myelogenous leukaemia patients in north India. Indian J Med Res. 2012;135:42-8.
- [Google Scholar]
- BCR-ABL mutations in chronic myeloid leukaemia. Hematol Oncol Clin North Am. 2011;25:997-1008.
- [Google Scholar]
- The breakpoint cluster region gene on chromosome 22q11 is associated with bipolar disorder. Biol Psychiatry. 2005;57:1097-102.
- [Google Scholar]
- Bipolar disorder associated with interferon-alpha treatment. Postgrad Med J. 1997;73:834-5.
- [Google Scholar]
- Candidate single nucleotide polymorphism markers for arsenic responsiveness of protein targets. Bioinform Biol Insights. 2010;4:99-111.
- [Google Scholar]






