Translate this page into:
An alternative strategy to generate coding sequence of macrophage migration inhibitory factor-2 of Wuchereria bancrofti
Reprint requests: Dr S.L. Hoti, Division of Microbiology & Immunology, Vector Control Research Centre (ICMR), Indira Nagar, Medical Complex, Puducherry 605 006, India e-mail: slhoti@yahoo.com
-
Received: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Different developmental stages of Wuchereria bancrofti, the major causal organism of lymphatic filariasis (LF), are difficult to obtain. Beside this limitation, to obtain complete coding sequence (CDS) of a gene one has to isolate mRNA and perform subsequent cDNA synthesis which is laborious and not successful at times. In this study, an alternative strategy employing polymerase chain reaction (PCR) was optimized and validated, to generate CDS of Macrophage migration Inhibitory Factor-2 (wbMIF-2), a gene expressed in the transition stage between L3 to L4.
Methods:
The genomic DNA of W. bancrofti microfilariae was extracted and used to amplify the full length wbMIF-2 gene (4.275 kb). This amplified product was used as a template for amplifying the exons separately, using the overlapping primers, which were then assembled through another round of PCR.
Results:
A simple strategy was developed based on PCR, which is used routinely in molecular biology laboratories. The amplified CDS of 363 bp of wbMIF-2 generated using genomic DNA splicing technique was devoid of any intronic sequence.
Interpretation & conclusions:
The cDNA of wbMIF-2 gene was successfully amplified from genomic DNA of microfilarial stage of W. bancrofti thus circumventing the use of inaccessible L3-L4 transitional stage of this parasite. This strategy is useful for generating CDS of genes from parasites that have restricted availability.
Keywords
Coding sequence
exon assembly
filarial genome database
macrophage migration inhibitory factor
polymerase chain reaction
Wuchereria bancrofti
Human lymphatic filariasis (LF) is a parasitic disease caused mainly by lymphatic filarial parasite Wuchereria bancrofti and partly by Brugia malayi and B. timori1. The disease is prevalent mainly in tropical and sub-tropical countries. The Global Programme to Eliminate Lymphatic Filariasis (GPELF) was launched in 2000 and after completing 13 years has resulted in substantial reduction in LF burden2. Research on lymphatic filariasis is mainly based on a zoonotic parasite, Brugia malayi as a model organism. The reason is the availability of an animal model Mastomys coucha to carry B. malayi infection and its developmental stages in laboratory3. This model provides a continuous source of DNA or proteins to study various molecular aspects of lymphatic filariasis. But, the source of developmental stages of W. bancrofti, a major causal organism of lymphatic filariasis, is not available for such studies. Beside this limitation, isolation of mRNA and generation of cDNA from certain stages of W. bancrofti is not possible due to difficulty in obtaining them4. Microfilariae of W. bancrofti can be obtained from carriers, but later stages (L1, L2 and L3) need to be raised in vectors which is not possible in all the laboratories, while other human stages are not accessible. Hence, obtaining mRNA from these stages to generate complete coding sequence (CDS) through reverse transcription (RT)-PCR is difficult. Further, conventional method of generating CDS requires mRNA isolation and RT-PCR kits, which are expensive, apart from the method being laborious and less successful at times. And, for successful amplification of a particular cDNA, high level of expression of the gene is often required. The presence of RNAses in the tissue samples decreases the success rate of cDNA amplification by RT-PCR. Genomic DNA splicing technique of generating cDNA directly from genomic DNA offers to circumvent these problems56. The data from recently ongoing genome sequencing project of W. bancrofti supported by the National Institute of Allergy and Infectious Disease, National Institutes of Health funded Genome Sequencing Centre for Infectious Diseases at the Broad Institute (http://www.broadinstitute.org) offer an opportunity to generate cDNA of different genes directly from genomic DNA. In the present study, the cDNA of Macrophage migration Inhibitory Factor-2 (wbMIF-2), a gene expressed in the inaccessible molting stage (from L3 to L4) of W. bancrofti7, was amplified using genomic DNA of microfilarial stage. The gene was identified from Filarial Genome Project sequence database (http://www.broadinstitute.org/) and its CDS was amplified directly from genomic DNA by using a simple genomic exon splicing strategy. Amplified coding sequence of wbMIF-2 can be further used for cloning, expression and functional characterization.
Material & Methods
The study was conducted in the Division of Microbiology and Immunology of Vector Control Research Centre (ICMR), Puducherry, India.
Gene scanning and annotation: Genome sequence of W. bancrofti was obtained from the Filarial Worms Sequencing Project, Broad Institute of Harvard and Massachusetts Institute of Technology (MIT), USA (http://www.broadinstitute.org/). The gene was identified and annotated using MEGA 5 software8 by alignment of the cDNA sequence of Macrophage migration Inhibitory Factor-2 of B. malayi (BmMIF-2, AY004865.1), obtained from National Centre for Biotechnology Information (NCBI) databank (http://www.ncbi.nlm.nih.gov/).
Designing of primers: Four primers were designed for the amplification of wbMIF-2 using Gene runner (http://www.generunner.net/) by optimizing melting temperature (Tm) and loop formation parameters. One set of primers (MIF-2F1 & MIF-2R1) was designed for the amplification of full length of wbMIF-2 gene and another set of overlapping primers (MIF-2OF1& MIF-2OR1) designed on the basis of exonic sequences. The primer sequences are: MIF-2F1: 5’- ATG CCG CTG ATA ACG CTT G- 3’; MIF-2R1: 5’- TTA TTT CTT CAT AAG CTC TTT CAT TGT - 3’; MIF-2OF1: 5’-TTA CAG TGG TTA AAT CGA TTG GAT C-3’; MIF-2OR1: 5’-CAC TGT AAG ACA TGA TGG ATT TTC A-3’.
PCR amplification of full gene and exons: The full length wbMIF-2 gene of 4.275 kb was amplified using genomic DNA extracted from microfilariae (n=50) of W. bancrofti, purified from blood9. The microfilaraemic blood (0.5 ml) collected at night from a carrier residing in Puducherry was used for DNA extraction. The DNA extraction was carried out using QIamp DNA micro kit (Qiagen, Germany) and the concentration was estimated in GenQuant (Amersham Bioscience, USA). The 50 µl of PCR reaction mix contained 1.5 µl of 10pmol of each primer (MIF-2F1 and MIF-2R1), 2x of PrimeSTAR Max master mix (TAKARA, Japan) and 50 ng of genomic DNA template. The PCR cycle parameters were as follows: initial denaturation at 98°C for four min followed by 35 cycles of denaturation at 98°C for 10 sec, annealing at 50°C for 30 sec, extension at 72°C for two min with final extension at 72°C for 10 min. The amplified full length wbMIF-2 PCR product was run on 0.8 per cent agarose gel with Ethidium bromide and observed under UV light. The wbMIF-2 band of 4.275 kb was cut and purified by using a nucleic acid gel purification kit (Macherey-Nagel, Germany). This purified product was used as a template for amplification of the two exons, in separate reactions. The overlapping primers were designed within similar range of Tm to amplify both the exons using the same PCR cycle simultaneously. PCR parameters for amplification of both the exons were as follows: initial denaturation at 95°C for five min followed by 30 cycles of denaturation at 95°C for one min, annealing at 50°C for 20 sec, extension at 72°C for 30 sec, with final extension at 72°C for 10 min. The 50 µl of PCR reaction for amplification of the two exons contained 1 µl of 10 pmol of primers (MIF-2F1 and MIF-2OR1) and (MIF-2OF1 and MIF-2R1), 0.5 unit of PR polymerase (Merck, India), 20 mmol dNTPs (FinZyme, Finland) and 50 ng purified DNA template. The amplified exons were purified using gel extraction kit and used in the assembly PCR as templates for amplification of the CDS.
Assembly of exons in single PCR reaction: Both the amplified exons were joined together in assembly PCR. The PCR reaction mixture contained 1 µl of 10 pmol of each primer (F1 and R1), 0.5 units of PR polymerase (Merck, India), 20 mmol dNTPs (FinZyme, Finland) and 50 ng of both purified exons as a template. PCR cycle parameters were as follows: initial denaturation at 95°C for five min followed by 35 cycles of denaturation at 95°C for one min, annealing at 50°C for 20 sec and extension at 72°C for 30 sec, with final extension at 72°C for 10 min. The amplified full length of wbMIF-2 coding sequence was run on 1.5 per cent of agarose gel with ethidium bromide and visualized under UV.
Sequencing confirmation of amplified CDS: The identity of full length wbMIF-2 coding sequence generated was confirmed by sequencing in ABI 3130Xl sequencer (Applied Biosystem, USA). The sequencing reaction of 10 µl contained 4 µl of Big Dye terminator (Applied biosystem, USA), 2 µl of 1 pmol primer (F1) and 4 µl of nuclease free water. Cycle sequencing parameters were followed as per manufacturer instructions.
Results
The genomic exon splicing strategy is shown in the flow chart (Fig. 1). Identification of MIF-2 of W. bancrofti was done on the basis of B. malayi MIF-2 cDNA sequence available in NCBI databank, as W. bancrofti and B. malayi are closely related parasites and have very high similarity of gene sequences. The search based on B. malayi MIF-2 cDNA sequence in W. bancrofti genome sequence database using MEGA5 software, yielded full length wbMIF-2 gene of 4.275 kb in the contig ADBV01000282.1. Upon annotation of this, it was found that wbMIF-2 had two exons of 183 and 180 bp, interspersed with a very large intron of 3.912 kb (Fig. 2). Both the exons are separated by standard rule of GT-AG splicing sites present at the intron endings.
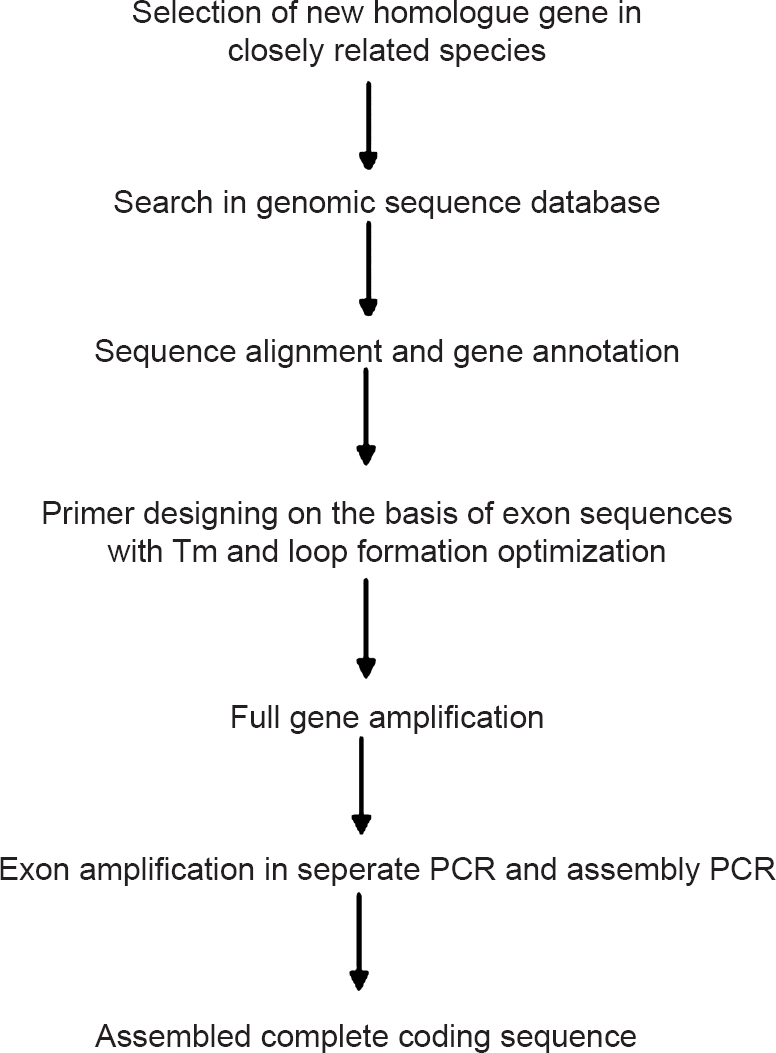
- Flow chart showing the complete process of genomic exon splicing strategy.

- Organization of W. bancrofti MIF-2 gene based on cDNA sequence and W. bancrofti contig ADBV01000282.1 from Filarial Genome Project sequences.
The complete nucleotide sequence of wbMIF-2 from W. bancrofti Genome Database was used for primer designing in Gene Runner software with optimization of Tm and loop formation (Fig. 3). The Tm of primers was set in the range of 2°C for each primer. Two sets of primers were designed for the amplification of full length wbMIF-2 gene and complete wbMIF-2 coding sequences. In the first PCR full length gene of wbMIF-2 of 4.275 kb was amplified (Fig. 4a), which was then used as a template for the amplification of the two exons (Fig. 4b) in PCR reaction, instead of using genomic DNA directly, to reduce the need for optimization efforts and enhancing the specificity. The strategy adopted by us required less genomic DNA for exon amplification compared to direct amplification of exons from the genomic DNA. Amplified exons were assembled in a separate PCR reaction using MIF-2F1 and MIF-2R1 primers and both the exons as template, which resulted in 363 bp complete coding sequence (Fig. 4c). The wbMIF-2 full coding amplicon was sequenced in 3031Xl sequencer and its identity was confirmed by aligning with W. bancrofti genome sequence. The sequence was found to match with a fragment between 12530 nt to 16804 nt, completely in the contig ADBV01000282.1.

- Complete coding sequence of Macrophage migration Inhibitory Factor-2 of W. bancrofti. Arrows indicate primer sequences designed on the basis of wbMIF-2 complete coding sequence.
-
Source: W. bancrofti genome database (http://www.broadinstitute.org).
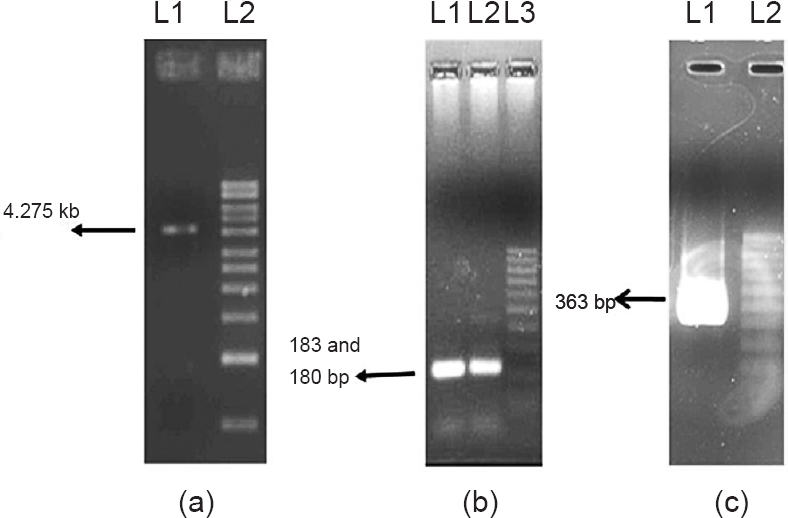
- PCR amplification of (a) MIF gene from genomic DNA of W. bancrofti. Lane 1, 4.275 kb amplicon, Lane 2- 1 kb marker; (b) MIF exons from MIF gene amplicon. Lane 1- 183 bp amplicon of first exon, Lane 2- 180 bp of amplicon of second exon, Lane 3- 100 bp ladder; and (c) Assembly of both exons in PCR. Lane 1- 363 bp assembled wbMIF-2 amplicon, Lane 2- 100 bp ladder.
Discussion
Parasitic agents remain a major cause of morbidity and mortality in humans in developing countries10 and lymphatic filarial parasite, W. bancrofti is one among them. Basic studies on the molecular aspects of developmental biology need to be continued to support the ongoing global LF elimination programme, especially to manage the morbidity. But, there are limitations to carry out such studies due to the scarcity of parasite material as certain developmental stages of the parasite are difficult to obtain or inaccessible, and a cyclic animal model is not available and there is no in vitro culture system for this parasite. Further, the expression of a particular gene may not be adequate to synthesize high amount of cDNA which limits its amplification in PCR, and isolation of mRNA and cDNA synthesis require much more expertise due to the presence of ubiquitous RNAase and less stability of mRNA101112.
Genome sequencing of W. bancrofti has created an opportunity to understand the mechanism of biological processes such as immune evasion by W. bancrofti. In the present study, we amplified the CDS of a gene from W. bancrofti, wbMIF-2 homologue, employing a modified strategy of genomic exon splicing technique which circumvented problem of cDNA synthesis from inaccessible molting L3 stage. In order to achieve this we identified wbMIF-2 homologue in the W. bancrofti genome database. The expression of wbMIF-2 homologue is specific to L3 molting stage (L3-L4) of W. bancrofti13 and considered as an immunomodulator at host-parasite interface14. The process of raising these stages of W. bancrofti is difficult, as it requires microfilaraemic blood sample, mosquito rearing facility and in vitro culture system. Use of DNA instead of RNA is always better in terms of simplicity, time consumption, reproducing results and cost-effectiveness. PCR based amplification of CDS of wbMIF-2 by using overlapping primers is easier and cost-effective compared to developmental stage maintenance, mRNA isolation and cDNA synthesis15. Previously, PCR based exonic assembly strategy has been used for the amplification of complete coding sequence of human genes with several modifications in primer designing and exon amplification16. However, in this method, human genomic DNA, which can be obtained in adequate quantities, was used directly as template in PCR reaction for amplification of each exon. But in the case of certain developmental stages of W. bancrofti the quantity of DNA available is very limited. To overcome this problem we amplified the full length gene of 4.275 kb and then used it as template for amplification of exons, which in turn were assembled in a separate PCR, to obtain CDS. Use of this strategy for amplification of complete coding sequence of a particular gene of W. bancrofti could solve the problem of isolation of mRNA with subsequent cDNA synthesis and need of high quantity of DNA from inaccessible parasitic stages. Further, this method will increase the specificity and reduce the optimization efforts in the amplification of complete coding sequence.
In conclusion, the strategy of genomic exon splicing is a simple, quick, cost-effective and a routinely used PCR based technique to amplify CDS of a particular gene from genomic DNA. Amplified coding sequence can be used for functional characterization of parasite genes and protein coded by such genes. In case of W. bancrofti, where certain developmental stages are not available for various studies, it can be a useful approach for generating CDS of various genes.
Acknowledgment
The authors acknowledge Dr P. Jambulingam, Director, Vector Control Research Centre (ICMR), Puducherry for his support. The financial support from Indian Council of Medical Research (ICMR), India is acknowledged.
Conflicts of Interest: None.
References
- Global programme to eliminate lymphatic filariasis: progress report for 2012. Wkly Epidemiol Rec. 2013;88:389-99.
- [Google Scholar]
- Progress and impact of 13 years of the Global Programme to Eliminate Lymphatic Filariasis on reducing the burden of filarial disease. PLoS Negl Trop Dis. 2014;8:e3319.
- [Google Scholar]
- Mastomys natalensis as an experimental host for Brugia malayi subperiodic. Southeast Asian J Trop Med Public Health. 1975;6:328-37.
- [Google Scholar]
- Immunological diagnosis of human filariases: present possibilities, difficulties and limitations. Acta Trop. 1974;31:108-28.
- [Google Scholar]
- SPLICE: a technique for generating in vitro spliced coding sequences from genomic DNA. Biotechniques. 2007;43:785-9.
- [Google Scholar]
- Rapid assembly of multiple-exon cDNA directly from genomic DNA. PLoS One. 2007;2:e1179.
- [Google Scholar]
- Brugia malayi excreted/secreted proteins at the host/parasite interface: stage-and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2009;3:e410.
- [Google Scholar]
- MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9.
- [Google Scholar]
- A simplified membrane filtration technique for the diagnosis of microfilaremia. J Parasitol. 1970;56:623-4.
- [Google Scholar]
- Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155-66.
- [Google Scholar]
- An alternative approach to synthesize cDNA bypassing traditional reverse transcription. Mol Biotechnol. 2008;39:201-6.
- [Google Scholar]
- Towards a new conceptual approach to “parasitoproteomics”. Trends Parasitol. 2005;21:162-8.
- [Google Scholar]
- The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol Biochem Parasitol. 2008;160:8-21.
- [Google Scholar]
- A new approach for cloning hLIF cDNA from genomic DNA isolated from the oral mucous membrane. Genet Mol Res. 2011;10:3455-62.
- [Google Scholar]






