Translate this page into:
Hybrid & El Tor variant biotypes of Vibrio cholerae O1 in Thailand
Reprint requests: Dr. Wanpen Chaicumpa, Emeritus Professor, Department of Parasitology, Faculty of Medicine Siriraj Hospital Mahidol University, Bangkok 10700, Thailand e-mail: tmwcc@mahidol.ac.th
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution Licence, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
El Tor Vibrio cholerae O1 carrying ctxBC trait, so-called El Tor variant that causes more severe symptoms than the prototype El Tor strain, first detected in Bangladesh was later shown to have emerged in India in 1992. Subsequently, similar V. cholerae strains were isolated in other countries in Asia and Africa. Thus, it was of interest to investigate the characteristics of V. cholerae O1 strains isolated chronologically (from 1986 to 2009) in Thailand.
Methods:
A total of 330 V. cholerae O1 Thailand strains from hospitalized patients with cholera isolated during 1986 to 2009 were subjected to conventional biotyping i.e., susceptibility to polymyxin B, chicken erythrocyte agglutination (CCA) and Voges-Proskauer (VP) test. The presence of ctxA, ctxB, zot, ace, toxR, tcpAC, tcpAE, hlyAC and hlyAE were examined by PCR. Mismatch amplification mutation assay (MAMA) - and conventional- PCRs were used for differentiating ctxB and rstR alleles.
Results:
All 330 strains carried the El Tor virulence gene signature. Among these, 266 strains were typical El Tor (resistant to 50 units of polymyxin B and positive for CCA and VP test) while 64 had mixed classical and El Tor phenotypes (hybrid biotype). Combined MAMA-PCR and the conventional biotyping methods revealed that 36 strains of 1986-1992 were either typical El Tor, hybrid, El Tor variant or unclassified biotype. The hybrid strains were present during 1986-2004. El Tor variant strains were found in 1992, the same year when the typical El Tor strains disappeared. All 294 strains of 1993-2009 carried ctxBC ; 237 were El Tor variant and 57 were hybrid.
Interpretation & conclusions:
In Thailand, hybrid V. cholerae O1 (mixed biotypes), was found since 1986. Circulating strains, however, are predominantly El Tor variant (El Tor biotype with ctxBC).
Keywords
Biotypes
El Tor variant
Thailand
Vibrio cholerae
Vibrio cholerae, the causative agent of severe watery diarrhoeal disease cholera, comprises 206 serogroups (O1-O206) based on antigenic diversity of their outer membrane lipopolysaccharides12. Strains of the O1 serogroup are divided into two biotypes i.e., classical and El Tor, according to their phenotypic differences. The classical strains are sensitive to 50 units of polymyxin B and Mukerjee’s type IV bacteriophage while the El Tor strains are generally dually resistant with the exception of some strains isolated in southern Bangladesh34. The El Tor strains are more adapted and resilient in environment, and cause higher infection to case ratio and more asymptomatic carriers than the classical counterpart5. Clinical manifestations of cholera caused by classical V. cholerae are more severe and prolonged than those caused by the El Tor67. This is attributable to the subtle difference of cholera toxin (CT) encoded by ctxAB genes of V. cholerae. Each of the V. cholerae O1 biotype can be divided into three serotypes i.e., Ogawa, Inaba, and Hikojima. Since 1817, the world has experienced seven cholera pandemics caused by V. cholerae O1. Strains of classical biotype were considered as the causative agents for the first six pandemics while the 7th cholera pandemic which started in 1961 from Sulawesi Island, Indonesia, was caused by El Tor V. cholerae O1. Since then, the El Tor V. cholerae had replaced the classical biotype as the sole cause of cholera epidemics until 1982 when there was a re-emergence of the classical V. cholerae isolated from patients during an epidemic in Bangladesh8–10. Both biotypes co-existed in Bangladesh until the classical vibrios became extinct in 1993. Until 1991, only toxigenic V. cholerae O1 strains caused cholera epidemic and pandemics. In 1992, a large cholera outbreak was reported from southern India and subsequently spread rapidly to neighbouring countries in several countries in Asia but did not spread to any other continent. The epidemic organism was non-O1 V. cholerae which could not be allocated into any of the pre-existing non-O1 serogroups. Subsequently, the organism was designated as serogroup O139 synonym Bengal in recognition of the place of origin11–13.
New V. cholerae O1 variants carrying mixed classical and El Tor phenotypes were first isolated from hospitalized patients with severe watery diarrhoea in Matlab, Bangladesh, in 20023. These isolates could not be allocated into the classical or El Tor biotype using conventional biotyping tests. Genotypically, these were found to carry the El Tor genome backbone including El Tor specific gene clusters: VSP-I and -II and RTX, indicating that these belonged to El Tor lineage. These isolates carried different combinations of alleles of tcpA and CTX prophage repressor gene (rstR)4. Their classical biotype characteristic was due to the presence of the classical CTX prophage and the deduced amino acids of the nucleotide sequence coding for cholera toxin B subunit belonged to classical biotype. Similar strains were isolated in Mozambique in 200414. Subsequently, V. cholerae O1 El Tor variants have been reported from several Asian countries including China, Japan, Hong Kong, Sri Lanka, and Vietnam and Africa (Zambia)15. In a retrospective study of V. cholerae strains isolated in Kolkata, India, during a 17 year period (1989-2005), using mis-match amplification mutation assay (MAMA)-PCR for determining ctxB alleles, it was revealed that the El Tor strains carrying ctxB allele of the classical biotype (ctxBC) have emerged since 1991 and co-existed with the prototype El Tor strains until 1995 when these completely replaced the typical El Tor biotype. Arbitrarily, the V. cholerae O1 strains carrying mixed phenotypes of classical and El Tor biotypes [polymyxin B (50 units) susceptibility and positive for chicken erythrocyte agglutination (CCA) and Voges-Proskauer (VP) test] are designated hybrid biotype where as the V. cholerae O1 with typical El Tor phenotypes (resistant to 50 units of polymyxin B, and positive for CCA and VP test) but carrying ctxBC are designated El Tor variant16. This nomenclature has been followed in this study.
The 7th pandemic cholera arrived in Thailand in 1963, when the El Tor strains completely replaced the classical vibrios and established endemicity17. The O139 Bengal was first isolated from hospitalized patient with severe watery diarrhoea in Thailand in 199318. The O139 serogroup completely disappeared from Thailand since 199617. Because it is known that classical V. cholerae strains with ctxBC inflicted more severe symptoms than the typical El Tor infection616 and because there had been a resurgence of cases of severe watery diarrhoea that required hospitalization during 1999-2002, it was of interest to make an insight into both phenotypic and genotypic characteristics of V. cholerae O1 isolated from cholera patients in different years in Thailand.
Material & Methods
Bacterial strains: A total of 330 V. cholerae O1 strains (248 Ogawa, 82 Inaba) isolated from hospitalized patients with cholera in various regions of Thailand from 1986 to 2009 (Table I) were investigated. Nineteen V. cholerae O1 strains collected from Australia, Bangladesh, India, Peru, Romania and Thailand in different years were used as reference strains419 (Table II). Among them, 16 strains were obtained from the collection of the Laboratory Science Division, the International Centre for Diarrhoeal Disease Research of Bangladesh, Dhaka, Bangladesh; two strains (G27875 and SC11) were provided by Dr T. Ramamurthy, the National Centre of Cholera and Enteric Diseases, Kolkata, India; and one strain (295/33) was from the Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand. All strains were subjected to conventional biotyping methods (susceptibility to 50 units of polymyxin B, CCA and VP test)20 using strains 569B and N16961 as the classical and El Tor reference strains, respectively.
| Year of isolation (n) | Strain no. | Serotype | Phenotype | Genotype | Biotype (see also Table IV) | Number of strain(s)/total number of strain(s) of the year | |||
|---|---|---|---|---|---|---|---|---|---|
| PB | CCA | VP | ctxB | rstR | |||||
| 1986 (5) | 1-2 | Inaba | R | + | + | E | E | El Tor | 2/5 |
| 3 | Inaba | R | + | + | E | E+C | El Tor | 1/5 | |
| 4 | Inaba | S | - | + | E | E | Hybrid group 1 | 1/5 | |
| 5 | Inaba | R | + | + | E+C | E | Unclassified group 1 | 1/5 | |
| 1987 (1) | 6 | Inaba | R | + | + | E | E | El Tor | 1/1 |
| 1989 (2) | 7 | Inaba | R | - | + | E+C | E+C | Hybrid group 2 | 1/2 |
| 8 | Inaba | S | + | + | E | E+C | Hybrid group 3 | 1/2 | |
| 1990 (13) | 9-12 | Inaba | R | + | + | E | E+C | El Tor | 4/13 |
| 13-16 | Ogawa | R | + | + | E | E | El Tor | 4/13 | |
| 17-18 | Inaba | R | + | + | E+C | E+C | Unclassified group 2 | 2/13 | |
| 19-21 | Ogawa | R | + | + | E+C | E | Unclassified group 1 | 3/13 | |
| 1991 (4) | 22 | Ogawa | R | + | + | E | E | El Tor | 1/4 |
| 23-25 | Ogawa | R | + | + | E+C | E | Unclassified group 1 | 3/4 | |
| 1992 (11) | 26 | Inaba | R | + | + | E | E+C | El Tor | 1/11 |
| 27 | Inaba | S | + | + | E | E | Hybrid group 4 | 1/11 | |
| 28 | Ogawa | R | + | + | E | E | El Tor | 1/11 | |
| 29 | Ogawa | R | + | + | E+C | E | Unclassified group 1 | 1/11 | |
| 30-33 | Ogawa | R | + | + | C | E+C | El Tor variant | 4/11 | |
| 34-36 | Ogawa | R | - | + | C | E+C | Hybrid group 5 | 3/11 | |
| 1993 (9) | 37-38 | Inaba | R | + | + | C | E+C | El Tor variant | 2/9 |
| 39-43 | Ogawa | R | + | + | C | E+C | El Tor variant | 5/9 | |
| 44 | Ogawa | R | + | - | C | E+C | Hybrid group 6 | 1/9 | |
| 45 | Ogawa | R | + | + | C | C | El Tor variant | 1/9 | |
| 1994 (7) | 46 | Inaba | R | + | - | C | E+C | Hybrid group 6 | 1/7 |
| 47-51 | Ogawa | R | + | + | C | E+C | El Tor variant | 5/7 | |
| 52 | Ogawa | S | + | + | C | E+C | Hybrid group 7 | 1/7 | |
| 1995 (11) | 53-62 | Ogawa | R | + | + | C | E+C | El Tor variant | 10/11 |
| 63 | Ogawa | R | + | - | C | E+C | Hybrid group 6 | 1/11 | |
| 1996 (3) | 64-65 | Ogawa | R | + | + | C | E+C | El Tor variant | 2/3 |
| 66 | Ogawa | S | + | + | C | E+C | Hybrid group 7 | 1/3 | |
| 1997 (3) | 67 | Ogawa | R | + | + | C | E+C | El Tor variant | 1/3 |
| 68-69 | Ogawa | R | + | + | C | C | El Tor variant | 2/3 | |
| 1998 (2) | 70-71 | Ogawa | R | + | + | C | C | El Tor variant | 2/2 |
| 1999 (179) | 72-78 | Inaba | R | + | + | C | C | El Tor variant | 7/179 |
| 79-83 | Ogawa | R | + | + | C | E+C | El Tor variant | 5/179 | |
| 84-85 | Ogawa | R | + | - | C | E+C | Hybrid group 6 | 2/179 | |
| 86-115 | Ogawa | R | + | - | C | C | Hybrid group 8 | 30/179 | |
| 116-247 | Ogawa | R | + | + | C | C | El Tor variant | 132/179 | |
| 248 | Ogawa | R | - | + | C | C | Hybrid group 9 | 1/179 | |
| 249 | Ogawa | R | - | - | C | C | Hybrid group 10 | 1/179 | |
| 250 | Ogawa | S | + | + | C | C | Hybrid group 11 | 1/179 | |
| 2000 (21) | 251-270 | Ogawa | R | + | + | C | C | El Tor variant | 20/21 |
| 271 | Ogawa | R | + | - | C | C | Hybrid group 8 | 1/21 | |
| 2001 (27) | 272-294 | Inaba | R | + | + | C | C | El Tor variant | 23/27 |
| 295-298 | Inaba | R | + | - | C | C | Hybrid group 8 | 4/27 | |
| 2002 (13) | 299-306 | Inaba | R | + | + | C | C | El Tor variant | 8/13 |
| 307 | Inaba | R | + | - | C | C | Hybrid group 8 | 1/13 | |
| 308-310 | Inaba | S | + | + | C | C | Hybrid group 11 | 3/13 | |
| 311 | Ogawa | R | + | + | C | C | El Tor variant | 1/13 | |
| 2003 (8) | 312-315 | Inaba | R | + | + | C | C | El Tor variant | 4/8 |
| 316 | Inaba | R | + | - | C | C | Hybrid group 8 | 1/8 | |
| 317 | Inaba | S | + | + | C | C | Hybrid group 11 | 1/8 | |
| 318 | Inaba | S | + | - | C | C | Hybrid group 12 | 1/8 | |
| 319 | Inaba | S | - | + | C | C | Hybrid group 13 | 1/8 | |
| 2004 (9) | 320-324 | Inaba | R | + | + | C | C | El Tor variant | 5/9 |
| 325-327 | Inaba | R | + | - | C | C | Hybrid group 8 | 3/9 | |
| 328 | Inaba | S | + | - | C | C | Hybrid group 12 | 1/9 | |
| 2009 (2) | 329-330 | Ogawa | R | + | + | C | C | El Tor variant | 2/2 |
n, total number of strain(s) of the indicated year; PB, susceptibility to 50 units of polymyxin B; CCA, chicken red blood cell agglutination; VP, Voges-Proskauer test; MAMA, mismatch amplification mutation assay; R, resistant; S, sensitive; +, positive; -, negative; C, classical; E, El Tor
| No. | Name of isolate (n=19) | Year of isolation | Country of origin | Serotype | Phenotype | Genotype | Biotype | Originally identified biotype | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PB | CCA | VP | ctxB | rstR | |||||||
| 1 | 569B | 1948 | India | Inaba | S | - | - | C | C | Classical | Classical |
| 2 | GP71 | 1971 | India | Ogawa | R | + | + | C | E | El Tor variant | El Tor |
| 3 | N16961 | 1975 | Bangladesh | Inaba | R | + | + | E | E | El Tor | El Tor |
| 4 | 2463-78 | 1978 | Australia | Inaba | R | - | - | C | C | Hybrid | El Tor |
| 5 | GP156 | 1979 | Australia | Ogawa | R | + | - | C | E | Hybrid | El Tor |
| 6 | 2164-88 | 1988 | United states | Inaba | R | + | + | C | C | El Tor variant | El Tor |
| 7 | 295/33 | 1990 | Thailand | Ogawa | R | - | + | E+C | E | Hybrid | El Tor |
| 8 | C6706 | 1991 | Peru | Inaba | R | + | + | E+C | E | Hybrid | El Tor |
| 9 | C7754 | 1991 | Romania | Ogawa | R | + | - | C | E+C | Hybrid | El Tor |
| 10 | MJ1485 | 1994 | Bangladesh | Inaba | R | - | + | C | C | Hybrid | El Tor |
| 11 | B33 | 2004 | Mozambique | Ogawa | R | + | - | C | C | Hybrid | El Tor |
| 12 | AR15493 | Unknown | Bangladesh | Inaba | R | + | + | C | E | El Tor variant | El Tor |
| 13 | AR15425 | Unknown | Bangladesh | Inaba | R | + | + | C | E | El Tor variant | El Tor |
| 14 | G27875 | Unknown | India (NICED) | Ogawa | R | + | + | C | E | El Tor variant | El Tor |
| 15 | SC11 | Unknown | India (NICED) | Ogawa | R | + | + | C | E | El Tor variant | El Tor |
| 16 | GP12 | Unknown | India | Ogawa | R | + | - | C | E | Hybrid | El Tor |
| 17 | AS230 | Unknown | India | Ogawa | R | + | + | C | E | El Tor variant | El Tor |
| 18 | AS231 | Unknown | India | Ogawa | R | + | + | C | E | El Tor variant | El Tor |
| 19 | AS233 | Unknown | India | Ogawa | R | - | + | C | E | Hybrid | El Tor |
PB, susceptibility to 50 units of polymyxin B; CCA, chicken red blood cell agglutination;, VP, Voges-Proskauer test; R, resistant; S, sensitive; +, positive; -, negative; C, classical; E, El Tor
Conventional- and MAMA-PCRs: All V. cholerae strains were examined for the presence of ctxA, ctxB, zot, ace, toxR, tcpAC, tcpAE, hlyAC and hlyAE by conventional PCR using strains AR15493 and AR15425 from Bangladesh as positive controls for zot, ace, toxR, and hlyA genes and strain C6706 as positive control for ctxAB and tcpA19. Conventional biotyping methods and a combination of MAMA- and conventional- PCRs were used for classifying the strains into prototype El Tor, hybrid, or El Tor variant biotypes, based on their ctxB and rstR genes21–23. Strains MJ1485 from Bangladesh and B33 from Mozambique served as hybrid biotype reference strains while G27875 and SC11 from NICED, India, were El Tor variant reference strains.
Primer sequences used in PCRs are shown in Table III19. Amplification mixture (25 μl) for ctxB-MAMA-PCR and rstR-PCR composed of 1 μl bacterial genomic DNA template, 2.5 μl 10× PCR buffer, 2 μl each of 2.5 mM deoxynucleotide triphosphate (Fermentas, Vilnius, Lithuania), 2 μl of 25 mM MgCl2, 2 μl of 10 μM of individual forward and reverse primers (Bio Basic Inc., Toronto, Canada), 0.5 units Taq DNA polymerase (Fermentas) and sterile ultra pure distilled water. Amplification of other genes was essentially the same as described previously19. The PCR products were analyzed by using 1.5 per cent agarose (Seakem LE, BMA, Glendate, CA, USA) gel electrophoresis and ethidium bromide staining (Sigma Chemical Co., USA). A Gel Doc 2000 (Bio-Rad, CA, USA) was used for DNA band documentation.
| Gene (s) | Primer sequence | Size of PCR amplicon (bp) | PCR condition | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Initial denaturation | Denaturation | Annealing | Extension | Final extension | No. of cycles | ||||
| Simple PCR | |||||||||
| rstRE | Forward: GCACCATGATTTAAGATGCTC | 501 (El Tor) | 94°C, 5 min | 94°C, 60 s | 58°C, 60 s | 72°C, 90 s | 72°C, 7 min | 30 | 22 |
| Reverse: TCGAGTTGTAATTCATCAAGAGTG | |||||||||
| rstRC | Forward: CTTCTCATCAGCAAAGCCTCCATC | 474 (Classical) | 94°C, 5 min | 94°C, 60 s | 64°C, 60 s | 72°C, 90 s | 72°C, 7 min | 30 | 22 |
| Reverse: TCGAGTTGTAATTCATCAAGAGTG | |||||||||
| MAMA-PCR | |||||||||
| ctxB | Forward: ACTATCTTCAGCATATGCACATGG | 96°C, 2 min | 96°C, 10 s | 55°C, 10 s | 72°C, 30 s | 72°C, 2 min | 25 | 21 | |
| Reverse for El Tor: CTGGTACTTCTACTTGAAACA | |||||||||
| Reverse for classical: CTGGTACTTCTACTTGAAACG | |||||||||
| MAMA-PCR, mismatch amplification mutation assay-PCR | |||||||||
Results & Discussion
All of the 330 V. cholerae O1 Thai clinical strains collected over 24 years (1986-2009) were found to carry ctxA, ctxB, zot, ace, toxR, tcpAE and hlyAE which verified genetically their toxin producing capacity and epidemic potential. Two hundred and sixty six strains were prototype El Tor (resistant to the polymyxin B, and positive for CCA and VP test) and the remaining 64 strains were not biotypable (Table I).
Identification of rstR by conventional PCR showed that the 36 strains of 1986-1992 carried either the El Tor rstR (rstRE/C) or combination of the El Tor and classical rstR (rstRE/C) (Table I). MAMA-PCR for ctxB of these isolates revealed that 18 (50%) carried ctxBE. Only 15 of these 18 strains had prototype El Tor phenotype (resistant to 50 units of polymyxin B, and positive for CCA and VP test) indicating that they were typical El Tor biotype. The other 3 strains, although carrying ctxBE, appeared to be hybrid biotype as they possessed mixed phenotypes (Table I and IV). There were 11 strains of 1986-1992 (31%) that carried ctxBE/C. Among these only one strain had mixed classical and El Tor phenotypes implying that this was hybrid biotype. The remaining 10 with ctxBE/C, however, could not be assigned into any of the redefined biotype scheme16 although these showed conventional El Tor phenotype (Tables I and IV. The remaining seven (19%) of the 1986-1992 (all were isolated in 1992) strains carried ctxB C; four of these had conventional El Tor phenotypes implying that these were El Tor variant while the other three had mixed phenotypes, and were hybrid (Table I). These data indicate the presence of hybrid biotype of V. cholerae O1 in Thailand since 1986 or even before and these co-existed with the typical El Tor strains. The V. cholerae O1 Thailand strains that carried ctxBE/rstREi.e., typical El Tor strains, were found for the last time in 1992 in this V. cholerae O1 collection which was the same year when the strains of El Tor variant biotype (strains 30-33) carrying ctxBC/rstRE/C emerged in the country (Table I). It is noteworthy that in 1992 the epidemic V. cholerae O139 strains emerged in Southern India11. The Fig. shows MAMA-PCR results of representative strains of V. cholerae chronologically isolated in Thailand i.e., ctxBC (Fig. A) and ctxBE (Fig. B).
| Biotype | Genotype | Phenotype | |||
|---|---|---|---|---|---|
| ctxB | rstR | PB | CCA | VP | |
| Classical | C | C | S | - | - |
| El Tor | E | E | R | + | + |
| El Tor | E | E+C | R | + | + |
| Hybrid group 1 | E | E | S | - | + |
| Hybrid group 2 | E+C | E+C | R | - | + |
| Hybrid group 3 | E | E+C | S | + | + |
| Hybrid group 4 | E | E | S | + | + |
| Hybrid group 5 | C | E+C | R | - | + |
| Hybrid group 6 | C | E+C | R | + | - |
| Hybrid group 7 | C | E+C | S | + | + |
| Hybrid group 8 | C | C | R | + | - |
| Hybrid group 9 | C | C | R | - | + |
| Hybrid group 10 | C | C | R | - | - |
| Hybrid group 11 | C | C | S | + | + |
| Hybrid group 12 | C | C | S | + | - |
| Hybrid group 13 | C | C | S | - | + |
| El Tor variant | C | C | R | + | + |
| El Tor variant | C | E+C | R | + | + |
| Unclassified group 1 | E+C | E | R | + | + |
| Unclassified group 2 | E+C | E+C | R | + | + |
PB, susceptibility to 50 units of polymyxin B; CCA, chicken red blood cell agglutination; VP, Voges-Proskauer test; R, resistant; S, sensitive; +, positive; -, negative; C, classical; E, El Tor
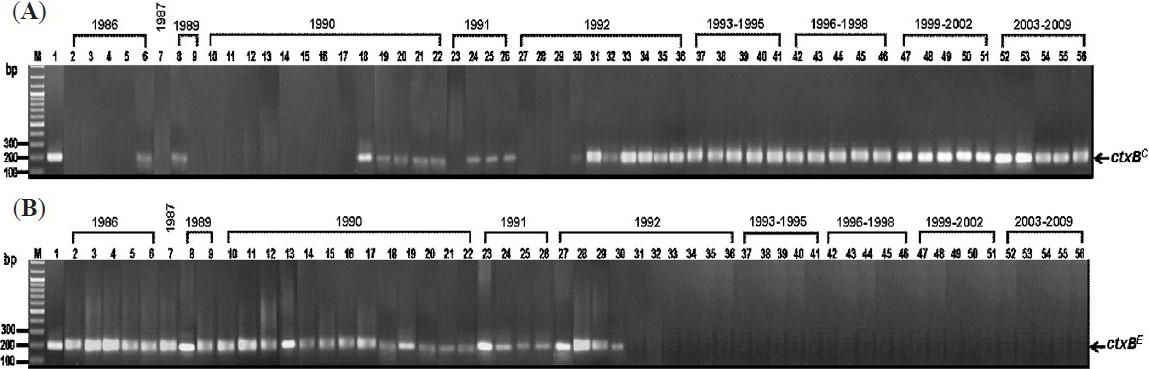
- Results of MAMA-PCR for amplification of ctxBC (A) and ctxBE (B) from representative V. cholerae strains isolated in Thailand during 1986-2009. Lanes 2-6, 1986 strains; lane 7, 1987 strains; lanes 8-9, 1989 strains; lanes 10-22, 1990 strains; lanes 23-26, 1991 strains; lanes 27-36, 1992 strains and lanes 37-56, 1993-2009 strains. Lane M, 100 bp DNA marker. Lane 1 in (A), positive control of ctxBC (569B); lane 1 in (B), positive control of ctxBE (N16961).
The V. cholerae O1 Thailand strains of 1993-2009 (294) were all found to carry ctxBC and either rstRC or rstR E/C. Majority of these strains (237 strains), however, were El Tor variants as their phenotypes were typical El Tor. The minority (57 strains) belonged to hybrid biotype because these had mixed phenotypes of classical and El Tor (Table I). The 1986-2009 Thailand strains with hybrid biotype could be arbitrarily classified into 13 different hybrid groups, 1-13 (Table IV). During 1986-1992, the biotypes of the 36 V. cholerae O1 Thailand strains were 15 prototype El Tor, 7 hybrid (groups 1-5), 4 El Tor variant, and 10 unclassified (unclassified groups 1 and 2) (Tables I and IV). The 294 strains of 1993-2009 belonged to hybrid groups 6-13 (57 strains) and El Tor variants (237 strains) (Tables I and IV).
The V. cholerae O1 of hybrid biotype was isolated from patients in India in 1991 when typical V. cholerae classical and El Tor biotypes co-existed suggesting the horizontal CTX prophage exchange between strains of the two principal biotypes in order for the infecting strains to be more adapted to the host hostile intestinal environment15 which conformed to the more severe cholera symptoms in the afflicted hosts in the recent years32224. It is noteworthy, however, that the classical V. cholerae O1 disappeared from Thailand since 196325 when the 7th cholera pandemic caused by typical El Tor strains first hit the Kingdom’s population. There has been no report on the period of co-existing classical and El Tor strains during 1986-2009 within Thailand. Our finding that the V. cholerae hybrid biotype could be detected among strains of 1986 suggested that there might be a re-emergence of the classical V. cholerae before or during 1986 or there might be other confounding molecular mechanism(s) in the shifting of the characteristics of V. cholerae bacteria in Thailand. The speculations warrant detail investigation. In 1992, the epidemic O139 strains emerged in India concurrent with the finding of El Tor variant in Thailand for the first time in this series of strain collection (Table I). Between 1992 and 1993, the V. cholerae O1 strains carrying ctxBC predominated in Kolkata, India15 and Thailand (this study). Thus, there seemed to be incomprehensible event of genetic evolution of the V. cholerae yielding strains of mixed traits/phenotypes of the two authentic biotypes during this period. After 1994, isolates of V. cholerae O1 in Kolkata, India, seemed to carry only ctxBC; thus these were El Tor variants or hybrids (no phenotypes were given to define the biotype)16. Similarity was found among the Thailand strains of this study, however, two years earlier than the Kolkata’s series. All of the Thai strains after 1992 carried ctxBC of which 57 (19%) were hybrid biotype and 237 strains (81%) were El Tor variants according to the conventional biotyping method and MAMA- and conventional- PCR determinations. In Punjab and Haryana, northern India, where a re-emergence of classical V. cholerae has not been reported, the V. cholerae hybrid biotype were also found in 2007 (80% of the isolates)26. As has been mentioned earlier, many V. cholerae isolates of several other countries in Asia and Africa were also found to be biotype hybrid/El Tor variant15 indicating that the El Tor V. cholerae bacteria, regardless of the geographical areas, tend to evolve for acquisition of the classical CTX prophage. This phenomenon will have impact, more or less, on the treatment of cholera, public health measures, as well as vaccine development.
The work was co-supported by the National Research University project of Thailand Office of Higher Education Commission (CHE) through Center for Biopharmaceutical Development and Innovative Therapy, Mahidol University and CHE RG 490329, the Thailand Research Fund (TRF; DPG5380001) and the Japan Health Science Foundation, Japan. P. Srimanote, N. Indrawattana, and N. Sookrung received research support from TRF.
References
- Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin, and addition of novel serogroups. Kansenshogaku Zasshi. 1997;71:1037-45.
- [Google Scholar]
- New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40:3296-9.
- [Google Scholar]
- Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. J Med Microbiol. 2006;55:1563-9.
- [Google Scholar]
- Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301-14.
- [Google Scholar]
- Seroepidemiologic studies during a simultaneous epidemic of infection with El Tor Ogawa and classical Inaba Vibrio cholerae. J Infect Dis. 1970;121(Suppl 121):S17-S24.
- [Google Scholar]
- History of cholera. In: Barua D, Greenough WB, eds. Cholera (3rd ed). New York: Plenum Medical Book Co; 1992. p. :1-36.
- [Google Scholar]
- Classical Vibrio cholerae biotype displaces El Tor in Bangladesh. Lancet. 1983;1:805-7.
- [Google Scholar]
- Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704.
- [Google Scholar]
- Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal Cholera Working Group. International Centre for Diarrhoeal Diseases Research, Bangadesh. Lancet. 1993;342:387-90.
- [Google Scholar]
- Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703-4.
- [Google Scholar]
- Mazambique Cholera Vaccine Demonstration Project Coordination Group. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J Med Microbiol. 2006;55:165-70.
- [Google Scholar]
- Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis. 2008;14:987-8.
- [Google Scholar]
- Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J Med Res. 2008;128:695-8.
- [Google Scholar]
- Ministry of Public Health, Thailand. Bureau of Epidemiology and the Department of Disease Control. Disease Notification Report 2000
- [Google Scholar]
- Virulence genes of clinical Vibrio cholerae O1 isolates in Thailand and their ribotypes. J Infect. 2007;55:557-65.
- [Google Scholar]
- World Health Organization, Geneva. Manual for laboratory investigations of acute enteric infections. WHO document CDD/83.3/Rev.1.113 1987
- [Google Scholar]
- Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype ElTor. Microbiol Immunol. 2008;52:314-7.
- [Google Scholar]
- Vibrio cholerae O1 clinical strains isolated in 1992 in Kolkata with progenitor traits of the 2004 Mozambique variant. J Med Microbiol. 2009;58:239-47.
- [Google Scholar]
- Emerging hybrid variants of Vibrio cholerae O1. In: Faruque SM, Nair GB, eds. Vibrio cholerae: Genomics and molecular biology. Norwich, UK: Horizon Scientific Press; 2008. p. :179-90.
- [Google Scholar]
- Molecular typing of Vibrio cholerae O1 isolates from Thailand by pulsed-field gel electrophoresis. J Health Popul Nutr. 2008;26:79-87.
- [Google Scholar]
- Department of Disease Control, Ministry of Public Health, no. ICD-10: A00. Bureau of Epidemiology, The Ministry of Public Health, Thailand. Available from: http://epi.moph.go.th/fact/Cholera.htm, accessed on January 10, 2010
- Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg Infect Dis. 2009;15:352-4.
- [Google Scholar]






