Translate this page into:
On the emergence of atypical Vibrio cholerae O1 El Tor & cholera epidemic
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Vibrio cholerae shows a remarkable variability easily seen by the identification of over 200 O serogroups in the species. However, only some lineages, of the O1 and O139 serogroups, have been able, so far, to trigger epidemic and pandemic cholera. The clinical O1 strains were further classified, on the basis of phenotypic and genotypic markers, into two biotypes: classical, which had driven, at least, the sixty cholera pandemic, and El Tor, the aetiological agent of the current cholera pandemic (7th cholera pandemic). The El Tor strains were first isolated on the Indonesian island of Sulawesi in 1961 and after that outbreaks, due to this lineage, started occurring in Bangladesh, India and Russia. Only more than a decade from its beginning the 7th cholera pandemic reached the North of Africa continent1.
By the end of the last century, two events changed centenary cholera concepts: the emergence of V. cholerae O139 serogroup2, causing epidemic cholera, and the discovery that the cholera toxin genes, which represent the major virulence factor in the cholera disease, are carried on the genome of a filamentous phage, CTXPhi3. The virulence, in the cholera syndrom, is multi-factorial, however, cholera toxin plays a key role in the severe secretory diarrhoea, that characterizes the devastating outbreaks. As being part of a phage genome the cholera toxin genes have an enourmous possibility of mobility by lateral gene transfer mechanism. In a short period of time it was determined that O139 serogroup was, in fact, an El Tor strain in a new robe4. This new V. cholerae phenotype was the consequence of changes in the O antigen-coding region but the overall traits and core genetic background of the El Tor biotype were maintained5. The expectation that this new serogroup would replace the major lineage driving the seventh cholera pandemic and would trigger the eighth cholera pandemic was not confirmed and, so far, V. cholerae O139 serogroup has been causing restricted outbreaks in the Indian subcontinent.
Cholera and V. cholerae continue challenge to the world and by the beginning of the 21st century a new scenario emerged when variants of typical V. cholerae El Tor biotype were identified, causing cholera outbreaks, in many Asian and African countries5–7. Interestingly, these variants harbour some phenotypic and genotypic traits that are characteristics of the two major pandemic V. cholerae biotypes. Classical and El Tor biotypes were successful in determining cholera pandemic during the last century, but the emerging atypical El Tor strains are likely to have a similar performance to these biotypes considering the epidemiology of these variants. Moreover, in some of the outbreaks, due to these atypical El Tor strains, a much higher proportion of patients presenting severe dehydration have been noticed. This is in contrast with what usually occurs when typical El Tor strains are in charge. In general, cholera produced by the classical biotype is more severe than that caused by El Tor, on the other hand, El Tor biotype presents an higher ecological fitness, with long persistence in the aquatic environment8. In the Fig. the current geographic distribution of these variants is represented. The black labelled countries are those where distinct atypical El Tor strains, the Matlab and Mozambique variants and the altered and hybrid El Tor, were characterized as determinants of outbreaks and, in some of these, there is no longer cholera outbreak due to typical El Tor lineage5. The hatched countries are those where one or more of the variants have been associated with cholera cases.
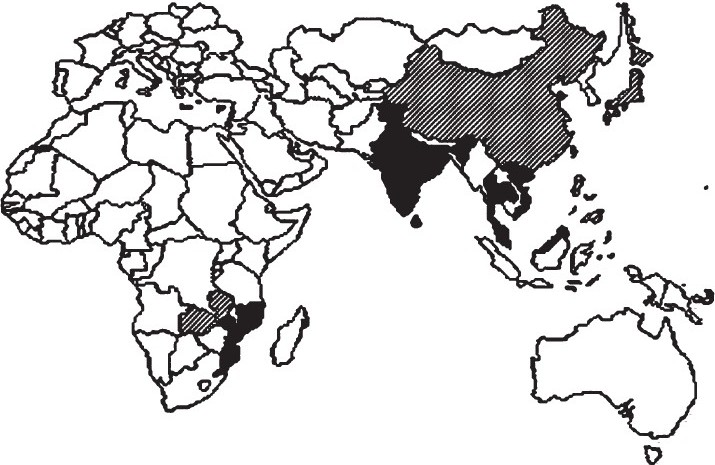
- Map showing the scenario of the spread of atypical V. cholerae. Countries where any atypical strain was the determinant of cholera outbreak (black label
 ) and countries that have already isolated atypical strains from cholera cases (hatched label
) and countries that have already isolated atypical strains from cholera cases (hatched label  ).
).Na-Ubol et al9, in this issue, enlarge this picture presenting a detailed genetic characterization of one representative collection of clinical V. cholerae O1 from Thailand recovered over 24 years (1986-2009). Their results showed the hybrid biotype of V. cholerae O1 strains co-circulating in Thailand, with the typical El Tor, since 1986. Considering their collection, the typical El Tor strains were found for the last time in 1992, when the El Tor variant biotype carrying ctxB C/rstRE/C emerged in the country. During 1993-2009 the majority of the strains characterized belonged to El Tor variants group including the hybrid biotype that harbours the classical cholera toxin allele ctxB. Therefore, Thailand is one of the Asian countries where the typical El Tor strains were displaced by the variants.
All the evidences on the shifting in the prevalence of the major V. cholerae El Tor lineage, as determinant of cholera epidemic, stress the urgency to perform genetic characterization of clinical V. cholerae isolates from the ongoing, as well previous cholera outbreaks.
The current scenario of the distribution in the world, of the atypical El Tor strains causing cholera epidemics, reveals that distinct pathogenic strains harbouring a combination of biotype traits, that can be genes coding for both virulence and ecological fitness, are likely to be successful as important public health threats.
References
- Cholera. In: Barua D, Greenough WB, eds. Cholera (3rd ed). New York: Plenum Medical Book Co; 1992. p. :1-36.
- [Google Scholar]
- Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet. 1993;341:1346-7.
- [Google Scholar]
- Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910-4.
- [Google Scholar]
- Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis. 2008;14:987-8.
- [Google Scholar]
- Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301-14.
- [Google Scholar]
- Hybrid & El Tor variant biotypes of Vibrio cholerae O1 in Thailand. Indian J Med Res. 2011;133:387-94.
- [Google Scholar]





