Translate this page into:
Antimicrobials & cholera: are we stranded?
Reprint requests: Dr. Amit Ghosh, National Institute of Cholera & Enteric Diseases, P-33 CIT Road, Scheme XM, Beliaghata, Kolkata 700 010, India e-mail: amitghosh24@yahoo.com
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Antimicrobial resistance poses a major threat in the treatment of infectious diseases. Though significant progress in the management of diarrhoeal diseases has been achieved by improved hygiene, development of new antimicrobials and vaccines, the burden remains the same, especially in children below 5 yr of age. In the case of cholera, though oral rehydration treatment is the mainstay, antimicrobial therapy is mandatory at times to reduce the volume of stool and shorten the duration of the disease. Though for many pathogens, antimicrobial resistance emerged soon after the introduction of antibiotics, Vibrio cholerae remained sensitive to most of the antibiotics for quite a long period. However, the scenario changed over the years and today, V. cholerae strains isolated world over are resistant to multiple antibiotics. A myriad number of mechanisms underlie this phenomenon. These include production of extended-spectrum beta-lactamases, enhanced multi-drug efflux pump activity, plasmid-mediated quinolone and fluoroquinolone resistance, and chromosomal mutations. Horizontal transfer of resistance determinants with mobile genetic elements like integrons and the integrating conjugative elements (ICEs), SXTs help in the dissemination of drug resistance. Though all strains isolated are not resistant to all antibiotics and we are not as yet “stranded”, expanding spectrum of drug resistance is a definite cause for concern. Pipelines of discovery of new antibiotics are drying up as major pharmaceutical companies are losing interest in investing money in this endeavour, mainly due to the short shelf-life of the antibiotics and also due to the fast emergence of drug resistance. To address this issue, attempts are now being made to discover drugs which are pathogen specific and target their “virulence mechanisms”. It is expected that development of resistance against such antibiotics would take much longer. This review briefly focuses on all these issues.
Keywords
Cholera
genetic elements
multidrug resistance
resistance genes
V. cholerae
Introduction
Discovery of effective agents to prevent and treat infections caused by pathogenic microorganisms has been one of the hallmarks of modern medicine. Antimicrobial agents are categorized according to their mechanism of action that include interference with cell wall synthesis (e.g., beta-lactams and glycopeptide agents), inhibition of protein synthesis (macrolides and tetracyclines), interference with nucleic acid synthesis (fluoroquinolones and rifampin), inhibition of metabolic pathways (trimethoprim-sulphamethoxazole), and disruption of bacterial membrane structure (polymyxins and daptomycin). Due to the excessive use of antimicrobials, treatment of bacteria-mediated diarrhoeal infections are becoming complicated, as many bacteria have become resistant to antimicrobial agents. The mechanism of resistance could be many; bacteria may be intrinsically resistant to these antimicrobial agents, or may acquire resistance through mutation or via the acquisition of resistance genes from other organisms. Acquired resistance genes may enable a bacterium to produce enzymes that destroy the antibacterial drug, to express efflux systems that prevent the drug from reaching its intracellular target, to modify the drug’s target site, or to produce an alternative metabolic pathway that bypasses the site or pathways of the action of the drug. Acquisition of genetic material(s) by antimicrobial-susceptible bacteria from resistant strains may occur through conjugation, transformation, or transduction, with transposons that facilitates incorporation of the resistance genes into the host’s genome or plasmids.
Soon after the discovery of antibiotics and their very successful application in medicine, antibiotic resistance started appearing in many microbes, however Vibrio cholerae continued to remain sensitive for a long period. In a worldwide survey carried out in 1976, only 3 per cent of the randomly collected strains were found to be resistant to commonly used antibiotics1. This scenario however, changed rapidly primarily due to the indiscriminate use of antibiotics. In a survey conducted in Bangladesh three years later, about 18 per cent of the “surveyed” strains were found to be resistant to a number of commonly used drugs2.
Pattern of antimicrobial resistance in V. cholerae is changing rapidly and some features are becoming well established. Based on several published works from National Institute of Cholera and Enteric Diseases, Kolkata, changing profile of resistance in V. cholerae is shown (Fig.). This is not unique as many investigations conducted in different geographical areas have demonstrated an increase in the antimicrobial resistance spectrum among epidemically significant V. cholerae over time.
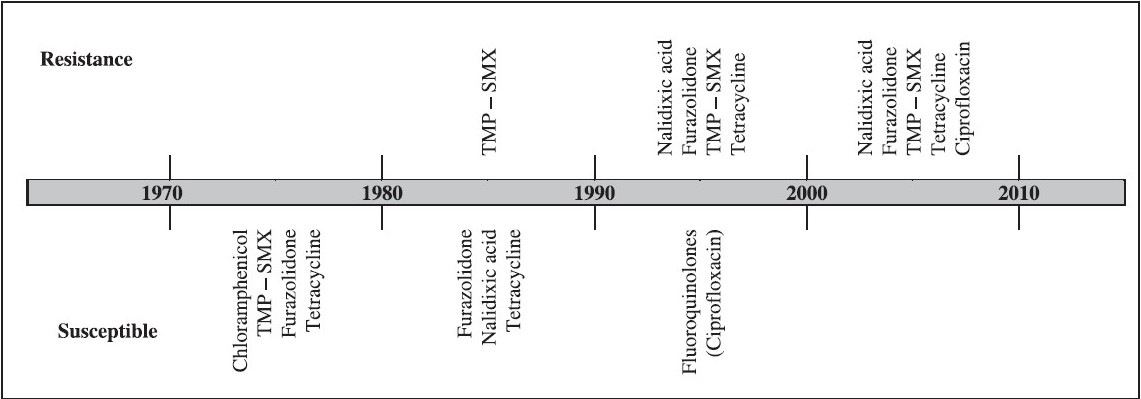
- Trends in antimicrobial resistance of V. cholerae in Kolkata, India extending a span of 40 years from 1970. (Information was collected from various published reports and Annual reports of NICED, Kolkata).
Furazolidone (Furoxone) was extensively used during late 1950s for the specific and symptomatic treatment of bacterial or protozoal diarrhea3. This antibiotic was recommended for the treatment in children. However, from late 1980s, almost all the enteric pathogens developed resistance to this drug and though its application is now very limited, this trend is still continuing. Analysis of the antibiograms of V. cholerae O1 from various regions of the world between 1938 and 1993 showed resistance to 1 to 3 antimicrobials, whereas the strains isolated from 1994 to 2005 possessed 3 to 8 resistance markers including fluoroquinolones.
Strategies adopted by V. cholerae to combat antimicrobials
(i) Resistance for quinolone and fluoroquinolone
In India and other countries, emergence of fluoroquinolone resistance has been reported from early 2000s as this drug was in extensive use for the treatment of different infectious diseases, including cholera. In many bacteria, resistance to the quinolones is generally associated with amino acid substitutions in portions of GyrA and ParC proteins called quinolone resistance-determining regions (QRDRs). Quinolone resistance in vibrios is mainly due to the occurrence of mutation in gyrA, which encodes a subunit of DNA gyrase, followed by mutations in parC, which encodes a subunit of DNA topoisomerase IV. In addition to the mutations in gyrA and parC, proton motive force-dependent efflux is also involved in quinolone resistance in clinical isolates of V. cholera4. Extensive use of ciprofloxacin has compromised its effectiveness in many countries due to the decreasing susceptibility of V. cholerae.
(ii) Integrons
Integrons are naturally occurring gene acquisition systems which “help” bacteria capture exogeneous genes and incorporate them into their genomes56. Integrons consist of a gene intI encoding a site-specific recombinase belonging to tyrosine-recombinase family called “integrase”, a recombination site att1 into which the exogenous gene cassettes, harbouring the recombination site attC, are inserted through site-specific recombination and a promoter Pc, located within intI that drives transcription of the captured gene. Integrons play a prominent role in the dissemination of drug resistance because these frequently carry drug resistance genes and are often associated with mobile genetic elements. Based on the sequence of intI, integrons have been divided into five classes. Out of these, class 1 integrons are found widely among the clinical isolates of pathogenic bacteria. IntI integrase of class 1 integrons possesses all the features needed for performing recombination between att1 and attC, however, the rate of cassette recombination is controlled by the transcriptional repressor LexA7. Till date more than 100 different types of integron-borne gene cassettes, most of which code for antibiotic resistance, have been discovered. Out of these, only a limited few have been found in V. cholerae even though integrons have been detected in a large number of V. cholerae strains isolated all over the world8. Besides the five classes of integrons alluded to above, there exists another class called “Superintegrons”. These are chromosomally located and are sedentary in the sense that these do not move. A superintegron was first discovered in V. cholerae but are now known to be present in many g-protobacteria5. Superintegrons harbour hundreds of genes but the functions of most of which are unknown. In V. cholerae O1, a few display significant homology to a number of drug resistance genes, suggesting that under appropriate conditions these could “become” drug resistance genes and confer upon its host the ability to express resistance phenotype. It may be mentioned here that in two strains of V. cholerae O1, one isolated in Brazil and the other in Vietnam, an integron borne qnr gene responsible for resistance to ciprofloxacin, have been detected910.
(iii) Integrative and conjugative element (ICE)
The newly discovered integrating conjugative elements are movable linear DNA elements, which can integrate into bacterial genome and “move” through conjugation. These have the capacity to incorporate genes encoding many functions from drug-resistance to DNA repair pathways11. SXT in V. cholerae is an ICE, which carries resistance genes for sulphamethoxazole-trimethoprim, streptomycin and chloramphenicol11. SXT exconjugants may contain tandem SXT arrays and promote the formation of novel ICEs12. The array formation appears to depend on conjugative transfer and is recA-independent. SXT excises from the chromosome to form a circular but non-replicative extrachromosomal molecule that is required for its transfer. The IncJ elements such as R391 are now found to be closely related to SXT11. Till date, more than 25 members of the SXT/R391 family of ICEs have been identified in environmental and clinical isolates of diverse species of gamma-proteobacteria worldwide.
The 100-kb ICE was first identified in a V. cholerae O139 clinical strain isolated in 1992 in India. Prior to the emergence of V. cholerae O139, SXT was rarely detected in O1 serogroup13. After the emergence of the serogroup O139, however, ICEs began to be detected frequently in Asian V. cholerae strains1113. Even among O139 strains, the SXT elements are not stably maintained and this results in periodic change in the strains in their resistance to sulphamethoxazole-trimethoprim, chloramphenicol, and streptomycin. SXT or closely related ICEs are now being reported in most clinical and some environmental strains of V. cholerae isolated in Asia and Africa. An SXT-related ICE, ICEVchMex1 identified in a Mexican environmental V. cholerae isolate provided the first description of an SXT-related ICE in the Western Hemisphere14. The significant difference between the SXT and ICEVchMex1 suggests that these ICEs evolved independently.
It was discovered by Beaber et al15 that ‘SOS response’ can enhance the conjugative transfer of SXT by promoting the expression of genes necessary for the transfer of SXT through the inactivation of SetR, an SXT encoded repressor that keeps the transfer genes repressed. Ciprofloxacin which can induce SOS response was found to be able to promote the transfer of SXT and thus facilitate its dissemination. In other words, it was found that therapeutic agents too can aid in the spread of antibiotic resistance. Kim et al10 found that ciprofloxacin activity is further compromised in strains habouring qnrVC3, which encodes a pentapeptide repeat protein of the Qnr subfamily as this protein protects topoisomerases from quinolone action. The gene qnrVC3 is present within a member of the SXT integrating conjugative element family found commonly on the chromosomes of multidrug-resistant strains of V. cholerae.
(iv) Extended-spectrum beta-lactamases
Beta-lactamase production could be demonstrated in 92.8 per cent of the ampicillin resistant V. cholerae strains isolated in south India16. The CTX-M-ases, which hydrolyze cefotaxime efficiently, are mostly encoded by transferable plasmids, and the enzymes have been found predominantly in many members of Enterobacteriaceae and V. cholera17. The CTX-M-ases belong to the molecular class-A beta-lactamases, and the enzymes are functionally characterized as extended-spectrum beta-lactamases which confer resistance to penicillin, extended-spectrum cephalosporins, and monobactams. Class I integron-associated orf513 also seems to be involved in the mobilization of blaCTX-M genes. CTX-M-type, PER-2-type and TEM-1-like enzymes were identified in V. cholerae strains isolated from cholera cases in Argentina18. Plasmid profile analysis and Southern blotting revealed the presence of plasmids of about 150 kb with genes encoding CTX-M-type or PER-2-type ESBLs.
(v) Plasmids
One of the common modes of the dissemination of drug resistance is through the plasmids. V. cholerae O1 strains isolated before 1970s were susceptible to tetracycline. Subsequently, due to extensive use of this drug, resistance to tetracycline was reported in many African countries. Strains isolated in Angola co-transferred tetracycline and chloramphenicol phenotype to E. coli. Plasmids belonging to incompatibility group (Inc) C and J were detected in V. cholera1920. Some of the V. cholerae O139 from India showed multidrug resistance. These strains harboured a 200 kb self-transmissible plasmid that mediated resistance to tetracycline, ampicillin, chloramphenicol, kanamycin, gentamicin, sulphamethoxazole and trimethoprim21. V. cholerae O1 El Tor isolated from patients in Uganda possessed a 130-MDa plasmid of incompatibility group 6-C that conferred resistance to trimethoprim (mediated by a dfrI gene), sulphonamides (suII), tetracycline (tetC), chloramphenicol (catI), ampicillin (a beta-lactamase gene different fromblaTEMor blaSHV), and streptomycin22. This plasmid was transferred from V. cholerae to many other enteric bacteria indicating its potential to spread.
The emergence of pMRV150-like or pIP1202-like plasmids in many bacterial pathogens and non-pathogens in many geographical areas pose an increasing health risk, as these mediate antimicrobial resistance to at least six antibiotics, ampicillin, streptomycin, gentamicin, tetracycline, chloramphenicol, and trimethoprim-sulphamethoxazole23. The plasmid pMRV150 was being found with increasing frequency in V. cholerae O139 from Hangzhou, eastern China from 1994 onwards and was found to be similar to the plasmid pIP1202, an IncA/C plasmid detected in an MDR Yersinia pestis isolated from a bubonic plague patient in Madagascar23.
(vi) Efflux systems
Bacteria may acquire efflux pumps that export antibacterial agents before it can reach its target site and exert its effect. In V. cholerae, efflux pumps responsible for resistance to many antimicrobials have been demonstrated. In many Gram-negative bacteria, resistance to antimicrobial agents is mediated by resistance-nodulation-division (RND) family efflux systems. Deletion of six genes encoding RND efflux pumps from the genome of the V. cholerae O1 El Tor strain N16961 made this strain sensitive to multiple antimicrobial compounds, including bile acids, antimicrobial peptides and antibiotics24. Colmer et al25 identified two open reading frames (ORFs) in V. cholerae with high degree of similarity with the conserved regions of the E. coli efflux pump proteins, EmrA and EmrB. NorM, a putative efflux pump of V. cholerae, is a member of the multidrug and toxic compound extrusion family of transporters. Singh et al26 demonstrated that NorM confers resistance to norfloxacin, ciprofloxacin, and ethidium bromide. When the “selected” amino acids in the periplasmic and cytoplasmic loops of NorM was mutated, V. cholerae became hypersensitive towards norfloxacin, there- by showing the importance of NorM in norfloxacin resistance.
V. cholerae chromosome contains many putative genes of the multidrug and toxic compound extrusion (MATE) family. These include vcrM vcmB, vcmD, vcmH and vcmN27. Elevated MICs of multiple antimicrobial agents, such as fluoroquinolones, aminoglycosides, ethidium, etc. were observed in a drug hypersusceptible strain of E. coli when these genes were introduced into it through transformation. It was further shown that efflux activities of VcmB, VcmD and VcmH were Na+-dependent. Besides these, an operon VccCAB consisting of three genes involved in multiple-drug resistance (MDR) through efflux has been indentified in V. cholerae. This operon is negatively regulated by a transcriptional autoregulatory protein belonging to the TetR family of transcriptional regulators28. As mentioned earlier, there are six operons of putative RND-type efflux transporters present in the chromosome of V. cholerae O1. Out of these, vexAB, vexCD or vexEF together with tolC(Vc) of V. cholerae NCTC4716 elevated MICs of various antimicrobial agents when introduced into a susceptible E. coli strain29. Though all these efflux pumps have been studied well, epidemiological significance of these in the dissemination of drug resistance remains unexplored.
The current conundrum
From the general survey that has been provided the moot point that emerges is that V. cholerae possesses a myriad variety of mechanisms to combat antibiotics and that a stage may soon come when the use of commonly used antibiotics may cease to become effective. Though it is not true that all strains everywhere are resistant to all antibiotics or there is no antibiotic available which could be effective against a recalcitrant strain, patterns of antibiograms of the recently emerging strains are a cause of great concern. If one examines the antibiotic “scenario” today, one finds that all antibiotics developed till date are targeted against only a few bacterial functions or enzymes and there too not all potential targets are targeted. For example, even though nineteen genes are known to be essential for DNA replication, only five are targeted.
This scenario, however, appears to be changing and “new” targets are being explored. Thus for example, in very recent times, a few promising lead molecules specific for V. cholerae have been obtained. One such molecule is Vibrepin30. In vibrios, replication of the smaller chromosome depends on rctB; Vibrepin, acts against V. cholerae by blocking RctB oriCII unwinding leading to the formation of large non-functional RctB complexes. Although Vibrepin appears to have targets other than RctB also, findings of Yamaichi et al30 suggest that RctB could be an attractive target for the generation of novel antibiotics specific to vibrios. The efflux pump inhibitors (EPIs) 1-(1-naphthylmethy1) piperazine (NMP) and phenylarginine-beta-naphthylamide (PAbetaN) are found to inhibit V. cholerae resistance-nodulation-division (RND) family efflux systems, hereby rendering the organism susceptible to antimicrobial agents. These were also found to inhibit the production of the virulence factors such as cholera toxin (CT) and the toxin coregulated pilus (TCP)31. Hence, one can think of developing RND efflux inhibitors as novel therapeutic agents for the treatment of cholera.
Thus in principle, there is considerable scope for the development of new antibiotics to combat drug resistance. In reality, however, it remains a doubtful proposition. Currently prevailing situation in the pharmaceutical industry makes it unlikely that a pharmaceutical company will spend millions in such an endeavour. A pharma company’s decision to invest in drug development is dictated by a parameter known as “Net Present value” (NPV)32. Any drug with an NPV less than 100 is usually not taken up for further development. NPV for an antibiotic is 100, whereas that for a muscular-skeletal drug is 115033. Antibiotics which are on the borderline is, therefore, are not attractive as investment targets. Indeed, it is seen that in the recent years there has been a sharp decline in the number of pharmaceutical companies engaged in research and development in the area of antibiotics.
Possible way out: Future directions
One of the major reasons why the investment in the research and development of new antibiotics is not an attractive financial proposition is the potentially limited clinical life span of antibiotics due to rapid development of resistance against them. Several measures to tackle this menace have been proposed- a prominent one being restricted use of antibiotics. However, a recent study has shown that this strategy may not be very effective in the long run. During a seven month period in 2001-2002 use of ciprofloxacin was restricted in Israel. Gottesman and his colleagues34 at Tel Aviv University measured ciprofloxacin sensitivity of E. coli isolated from urine, before, during and after this period. About 50 per cent reduction in ciprofloxacin use reduced the percentage of samples containing ciprofloxacin resistant bacteria from 12 to 9 per cent. But as soon as the restriction was lifted, a resurgence in the number of resistant bacteria occurred. Limited usefulness of this strategy pointed to the need for the exploration of other methods. A possible way could be to test an already developed antibiotic, untested against a particular pathogen, for its effectiveness against that pathogen. Feasibility of such an approach is demonstrated by the fact that antibiotic sitafloxacin, which has not been used in the treatment of cholera so far, has been found to be 4-6 fold more potent against V. cholerae, compared to ciprofloxacin, ofloxacin, sparfloxacin and levofloxacine in in vitro studies, suggesting that it can probably be used in the treatment of cholera caused by fluoroquinolone resistant strains of V. cholera35. This approach too does not provide a satisfactory solution as with time bugs will develop resistance against these antibiotics also. Therefore, to minimize the possibility of rapid development of drug resistance, an altogether different approach is being explored. Detailed knowledge of virulence mechanisms in many bacterial pathogens accumulated during the past two decades has led to the invention of a new class of antibiotics that target the virulence mechanism in such pathogens36. As these antivirulance drugs are pathogen-specific, their use are expected to be much more limited than the commonly used broad-spectrum antibiotics and therefore, it is expected that in the development of resistance against such antibiotics would take much longer37. Guided by such considerations and using a high-throughput phenotypic screen that inhibits virulence regulation, a small molecule 4- [n- (1,8-napthalimide]-n-butyric acid (designated Virstatin) has been identified38. By inhibiting the transcriptional regulator ToxT, Virstatin prevents expression of two critical virulence factors in V. cholerae namely, the cholera toxin (CT) and the toxin coregulated pilus (TCP) and thus renders it “avirulant”. Further, it has been seen that orogastric administration of virstatin protects infant mice from intestinal colonization of V. cholerae. Though the clinical efficacy of virstatin has not yet been explored, in principle at least it, opens up an avenue for the discovery of more such molecules.
Spread of antibiotic resistance has been recognized by the WHO as an extremely serious problem as it complicates the treatment of infectious diseases enormously. Therefore there is an urgent need to fight the spread of antibiotic resistance. If all the factors that have been mentioned are considered, it becomes apparent that perhaps a combination of conventional antibiotics and the antivirulence drugs, along with the “restricted” use of already available antibiotics, could turn out to be most effective method in arresting the explosive growth of drug resistance that is plaguing the world today. In this respect, several strategies were proposed at international and regional levels3940.
References
- Global surveillance of antibiotic sensitivity of Vibrio cholerae. Bull World Health Organ. 1976;54:181-5.
- [Google Scholar]
- Emergence of multiply antibiotic-resistant Vibrio cholerae in Bangladesh. J Infect Dis. 1980;142:939-42.
- [Google Scholar]
- Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agents Chemother. 2002;46:2676-8.
- [Google Scholar]
- A new in vitro strand transfer assay for monitoring bacterial class 1 integron recombinase IntI1 activity. PLoS One. 2007;2:e1315.
- [Google Scholar]
- Epidemiological and molecular aspects on cholera. In: Ramamurthy T, Bhattacharya SK, eds. Epidemiological and molecular aspects on cholera. Springer: New York; 2010. p. :291-310.
- [Google Scholar]
- New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg Infect Dis. 2008;14:1129-31.
- [Google Scholar]
- Transferable quinolone resistance in Vibrio cholerae. Antimicrob Agents Chemother. 2010;54:799-803.
- [Google Scholar]
- The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55:173-83.
- [Google Scholar]
- Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol. 2004;186:2636-45.
- [Google Scholar]
- Class 1 integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg Infect Dis. 2003;9:500-2.
- [Google Scholar]
- SXT-related integrating conjugative element in New World Vibrio cholerae. Appl Environ Microbiol. 2006;72:3054-7.
- [Google Scholar]
- SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72-4.
- [Google Scholar]
- Transferable plasmid-mediated drug resistance among non-O1 Vibrio cholerae and rough strains of Vibrio cholerae from Tamilnadu, India. J Hyg (Lond). 1984;92:59-65.
- [Google Scholar]
- Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum beta-lactamases. Can J Microbiol. 2004;50:137-65.
- [Google Scholar]
- Plasmidic extended-spectrum beta-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob Agents Chemother. 2002;46:1462-8.
- [Google Scholar]
- Antibiotic resistance of Vibrio cholerae: special consideration of R-plasmids. Zhonghua Min Guo Wei Sheng Wu Xue Za Zhi. 1978;11:99-103.
- [Google Scholar]
- Stability of R plasmids belonging to different incompatibility groups in Vibrio cholerae “El Tor”. Ann Mircobiol (Paris). 1978;129:409-14.
- [Google Scholar]
- Emergence of tetracycline resistance due to a multiple drug resistance plasmid in Vibrio cholerae O139. FEMS Immunol Med Microbiol. 1995;11:131-6.
- [Google Scholar]
- A transferable multiple drug resistance plasmid from Vibrio cholerae O1. Microb Drug Resist. 1995;1:203-10.
- [Google Scholar]
- Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob Agents Chemother. 2008;52:3829-36.
- [Google Scholar]
- Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun. 2008;76:3595-605.
- [Google Scholar]
- Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol Microbiol. 1998;27:63-72.
- [Google Scholar]
- Analysis of the topology of Vibrio cholerae NorM and identification of amino acid residues involved in norfloxacin resistance. Antimicrob Agents Chemother. 2006;50:3717-23.
- [Google Scholar]
- Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol Immunol. 2005;49:949-57.
- [Google Scholar]
- Characterization of the Vibrio cholerae vceCAB multiple-drug resistance efflux operon in Escherichia coli. J Bacteriol. 2005;187:5500-3.
- [Google Scholar]
- VceR regulates the vceCAB drug efflux pump operon of Vibrio cholerae by alternating between mutually exclusive conformations that bind either drugs or promoter DNA. J Mol Biol. 2005;349:387-400.
- [Google Scholar]
- Targeting the replication initiator of the second Vibrio chromosome: towards generation of vibrionaceae-specific antimicrobial agents. PLoS Pathog. 2009;5:e1000663.
- [Google Scholar]
- Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J Antimicrob Chemother. 2009;63:103-8.
- [Google Scholar]
- Impact of antibiotic restrictions: the pharmaceutical perspectives. Clin Microbiol Infect. 2006;12((Suppl 5)):25-34.
- [Google Scholar]
- Why is big pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427-30.
- [Google Scholar]
- Impact of quinolone restriction on resistance patterns of Escherichia coli isolated from urine by culture in a community setting. Clin Infect Dis. 2009;49:869-75.
- [Google Scholar]
- The potent antibacterial activity of Sitafloxacin against fluoroquinolone-resistant clinical isolates of Vibrio cholerae O1. Microbiol Immunol. 2007;51:467-9.
- [Google Scholar]
- Inhibitors of two-component signal transduction systems: inhibition of alginate gene activation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1993;90:965-9.
- [Google Scholar]
- Disarming pathogens - a new approach for antibiotic development. N Engl J Med. 2006;354:296-7.
- [Google Scholar]
- Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670-4.
- [Google Scholar]
- WHO Regional Strategy on Prevention and Containment and Antimicrobial resistance 2010-2015. Available from: http://www.searo.who.int/LinkFiles/BCT-htl-407.pdf, accessed on February 10, 2011
- [Google Scholar]
- The growing challenge of antimicrobial resistance in the South-East Asian Region - Are we losing the battle? Indian J Med Res. 2010;132:482-6.
- [Google Scholar]






