Translate this page into:
Physical state & copy number of high risk human papillomavirus type 16 DNA in progression of cervical cancer
Reprint requests: Dr Alok C. Bharti, Division of Molecular Oncology, Institute of Cytology & Preventive Oncology (ICMR), I-7, Sector-39, Noida, U.P.-201301, India email: bhartiac@icmr.org.in
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
High-risk human papilloma virus (HR-HPV) infection and its integration in host genome is a key event in malignant transformation of cervical cells. HPV16 being a dominant HR-HPV type, we undertook this study to analyze if viral load and physical state of the virus correlated with each other in the absence of other confounding variables and examined their potential as predictors of progressive cervical lesions.
Methods:
Both, viral load and integration status of HPV16 were determined by real time URR PCR and estimation of E2:E6 ratio in a total of 130 PGMY-RLB -confirmed, monotypic HPV16-infected cervical DNA samples from biopsies of cytology-confirmed low grade (LSIL, 30) and high grade (HSIL, 30), and invasive carcinoma, (squamous cell carcinoma SCC, 70) cases.
Results:
Investigation of DNA samples revealed a gradual increase in HPV16 viral load over several magnitudes and increased frequency of integration from LSIL to HSIL and HSIL to invasive cancer in relation to the severity of lesions in monotypic HPV16-infected cervical tissues. In a substantial number of precancer (11/60) and cancer cases (29/70), HPV16 was detected in concomitant mixed form. The concomitant form of HPV16 genome carried significantly higher viral load.
Interpretation & conclusions:
Overall, viral load and integration increased with disease severity and could be useful biomarkers in disease progression, at least, in HPV16-infected cervical pre-cancer and cancer lesions.
Keywords
Cervical cancer
copy number
HPV16E2
HSIL
human papillomavirus
LSIL
viral integration
viral load
Despite effective screening programmes and lately introduction of two successful prophylactic human papillomavirus (HPV) vaccines for cancer prevention, cervical cancer is still a leading gynaecological malignancy of the developing world1. Globally, there are approximately 530,000 new cases of cervical cancer with an annual mortality rate of 275,0002. Interestingly, of the total global cervical cancer burden, more than 85 per cent of the total cases and 88 per cent of the total mortality are contributed by developing countries. Among the high risk HPV types established as aetiological factors for cervical cancer, infection of HPV16 or HPV18 has collectively been implicated in causation of more than 70 per cent of global cervical cancer burden. In contrast to other regions of the world, upto 90 per cent of the cervical cancer lesions in Indian women harbour specifically the HPV type 16; whereas, presence of other HPVs including HPV18 is relatively low345. The dominance of HPV16 in comparison to other high risk HPV types particularly in global and Indian scenario indicates toward a major role of HPV16 infection. It is likely that factors such as viral copy number and physical state of viral genome (episomal vs. integrated or mix) may have important clinical implications in viral persistence and progression of cervical neoplasia as suggested in some earlier studies67. Integration is considered as a key event in cervical carcinogenesis which results in loss of episomal viral DNA as it is well-documented that E2 gene product derived from episomal DNA have an inhibitory effect on viral oncogene expression8, and integrants are spontaneously selected during cancer progression due to selective growth advantage and endogeneous antiviral response9. Therefore, apart from the expression of viral oncogenes E6 and E7 that lead to tumorigenic transformation, physical state of virus and viral copy number have been demonstrated to be the major risk factors for development of high-risk HPV-mediated cervical cancer610. On the contrary, integration of HPV16 viral DNA into the host genome and high viral copy number within infected epithelial cells have been associated with an increased persistence of HPV infection and an increased risk of developing cervical intraepithelial neoplasia 2/3 (CIN2/3) or cancer6711.
Unlike HPV18 which shows high frequency of integrated viral genomes in cervical carcinomas, only a proportion of cases ranging from 28 to 67 per cent, depending on the techniques used, demonstrate presence of integrated HPV16 in invasive cervical carcinoma1112. Some studies have demonstrated that integration of the HPV genome has also been found in low-grade lesions and even in normal cervices131415 whereas others have shown that not all invasive cancers carry the integrated HPV genome1416. However, besides a few sporadic reports417, data related to viral load and its physical status particularly with reference to HPV16- are lacking. The potential reason for higher pathogenicity of Indian HPV16 subtype may be related to its biological behaviour with respect to its viral load and integration events during progression of the early infection to cervical carcinogenesis. Moreover, investigations on the viral load and integration of HPV16 independently or in combination have been attempted in the past with variable results. The reasons for such variability may partly be ascribed to multiple infections which could have confounding effect. In the present study, we examined copy number and physical state of infecting monotypic high risk HPV16 in cervical cancerous and pre cancerous tissues.
Material & Methods
Clinical specimens and the study design: This cross-sectional study was performed on 130 prospectively collected and confirmed monotypic HPV16 positive tissue specimens comprising 70 malignant, 60 premalignant cervical tissues biopsies (colposcopy-guided). Pre-cancer cases were defined on cytology as low-grade (LSIL) or high-grade squamous intraepithelial lesions (HSIL). Samples were collected prior to any chemo/radio therapy from the Cancer Clinic, Gynaecology out patient department of Lok Nayak Hospital, New Delhi, India, during the period April 2008 to July 2011. DNA isolated from cervical tissues (biopsy samples) was analyzed for HPV16 viral load and physical state. Written informed consent was obtained from all the subjects included in the study and epidemiological details were taken from their clinical records. The study protocol was approved by the Institutional Ethics Committee of Institute of Cytology & Preventive Oncology (ICPO), New Delhi, India, where the laboratory work was carried out. The clinico-pathological characteristics are presented in Table I. A portion of each biopsy collected in cold 1X phosphate buffer saline (PBS) was immediately processed for molecular biological analysis and the other half was sent for histopathological diagnosis in formalin solution. All reagents used in the study were of analytical and molecular biology grade and were procured from Sigma Aldrich (USA) unless specified.
DNA extraction and diagnosis of HPV infection: Genomic DNA was isolated from precancer and cancer cervix biopsies by the standard phenol-chloroform and proteinase K digestion, and PCR amplification was performed following the procedure described earlier3. The quantity and quality of DNA was assessed using agarose gel electrophoresis and by UV spectrophotometry (Nanodrop, USA). The initial HPV diagnosis was performed by using a pair of L1 consensus degenerate primers (MY09 and MY11). HPV16 typing was done by type-specific primers318. PCR was performed in a 25µl reaction mixture containing 100ng DNA, 10mM Tris-HCl (pH 8.4), 50mM KCl, 1.5mM MgCl2, 125mM of each dNTPs (MBI fermentas, Ontario, Canada), 5pmol of oligonucleotide primers and 0.5U Taq DNA polymerase. β-globin gene was used as internal control. The temperature profile used for amplification constituted an initial denaturation at 95°C for 4 min followed by 30 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 1min, which was extended for 5min at the final cycle. Custom-synthesized, HPLC-purified primers were procured from M/s Microsynth (Germany). HPV16 positive samples were subjected to comprehensive HPV genotyping by PGMY-Reverse Line Blot which can detect about 32 HPV types including all high-risk and low-risk types19 and samples with monotypic HPV infection only were included in the study.
HPV16 viral load determination by real-time quantitative PCR (RQ-PCR): Quantification of HPV16 viral copy number and normalization for input cellular DNA copies was performed as described earlier20. Target-specific amplification of HPV16 URR (upstream regulatory region) and p53 exon5 were performed with a iQ-cycler system (Biorad, Hercules, USA) using a recommended iQSYBR green PCR supermix following the protocol provided by the manufacturer. Standard curves used to quantify HPV16 copy number were made with 10 fold serial dilutions of the WHO HPV16 international standard (NIBSC, London, UK) containing 50,000, 5000, 500, 50 and 5, HPV16 DNA copies diluted in background of C33a genomic DNA. Briefly, the reaction was performed in a final volume of 25μl containing 1X SYBR green super mix with 0.25μM of HPV16 URR forward and reverse primers and 50ng of genomic DNA of test samples. The URR primers were selected for viral load quantitation as these are retained in both episomal and integrated forms of the HPV16 viral genome. The PCR amplification was performed as follows: 1 cycle of 96°C for 3min, 40 cycle at 94°C for 30sec, 55°C for 30sec and 72°C for 30sec with realtime measurement performed during amplification step (72°C) at each cycle. Each realtime amplification was followed by melt curve analysis for confirmation of predicted amplicon20. Crude viral load or copy number of HPV16 genome in clinical sample was calculated by the interpolation of standard curves of the dilution series generated by the Sequence Detection Software (iCycler iQ software version 3.0) of iCycler iQ real time PCR detection system (Biorad, Hercules, USA). On the other hand, samples with viral loads higher than 50, 000 copies/reaction were diluted in water to bring it down to the range of standard curve. The viral load values were normalized to input host diploid genomic DNA using p53 exon5 amplification calibrated with C33a genomic DNA standard (NIBSC) as indicated below:
Normalized HPV16 viral load/Unit host cell genome = HPV16 URR copy number/ no. of diploid host genomes
The normalized HPV16 viral copy numbers are expressed as the number of viral copies/Unit host cell genome.
Determination of physical state of HPV16 genome in cervical tissues: To determine physical state of viral genome in HPV16 positive precancer and cancer tissues, full length primer for HPV16 E2 (validated by our laboratory previously)20, was utilized to analyze the presence of intact E2 ORF that is disrupted or deleted in integrated virus. Amplification of HPV16 E6, which is invariably retained in the integrated virus, was used as denominator of total HPV16 DNA irrespective of the physical state of the virus. Briefly, genomic DNA of HPV16-positive cases was used to assess the presence or absence of the HPV16 E2 gene, with reference to the HPV16 E6 gene. Genomic DNA of test samples (50ng) was PCR amplified for HPV16 E2 and E6 in a 25µl reaction mixture containing 10mM Tris-HCl (pH 8.4), 50mM KCl, 1.5mM MgCl2, 125µM of each dNTPs (dATP, dGTP, dCTP, dTTP), 5pmol of oligonucleotide primers for either full-length HPV16 E2 or HPV16 E6 and 0.5U AmpliTaqGold DNA polymerase (Applied Biosystems, USA). The amplification was performed with an initial denaturation at 95°C for 4 min, polymerization for 35 cycles of denaturation at 95°C for 30sec, annealing at 55°C for 30sec and extension at 72°C for 1min, which was extended for 5min at the final cycle (Applied Biosystems). Densitometric ratio of E2 and E6 amplicons was measured on AlphaDigiDoc using Alpha Ease FC version 4.1.0 (Alpha Innotech Corporation, USA) to determine the physical state of HPV16 for each sample. Densitometric ratios of E2:E6 amplicons of all clinical samples were normalized to E2:E6 ratio of vector-free HPV16 plasmid (a kind gift from Prof. H. zur Hausen, DKFZ, Germany) which was used as reference for pure episomal form of HPV16 genome and helped to normalize the variations in PCR efficiencies. DNA from SiHa cells (Procured from American Type Culture Collection, USA) was used as control for fully integrated DNA. The E2:E6 ratio in clinical samples with reference to the plasmid control was calculated by the following formula:
Normalized E2:E6 ratio of clinical samples = (IDV E2:IDV E6) samples /(IDV E2:IDV E6) plasmid
Where IDV indicates integrated densitometric values of DNA band of HPV16 E2 amplicon (IDVE2) or HPV16 E6 (IDVE6) amplicon of the plasmid and sample DNA. An E2:E6 ratio with a value of 0 represented completely integrated HPV16 genome and value of 1 or higher represented predominantly episomal viral genome, whereas values > 0 and < 1 indicated a mixed form of HPV16 DNA.
Statistical analysis: The data analysis was performed using the statistical software SPSS version 17 (SPSS Inc, USA) or Sigma Stat (Systat Software Inc., USA) or Graph Pad Instat (version 4.0) (Graph Pad Software, Inc., USA). Mann Whitney U test was used to compare the viral load and integration state of HPV16 between different disease states.
Results
To evaluate the status of HPV infection in terms of viral load and physical state of HPV16 genome in samples representative of the natural history of cervical cancer, DNA samples isolated from 130 monotypic HPV16 infection-confirmed cases comprising 60 pre-cancers (LSIL – 30; HSIL – 30) and 70 cancer cases for HPV16 viral load were analysed by real time quantitative PCR and physical state was determined by assessing densitometric ratio of HPV16 E2 and E6 gene amplification. The mean age of women in precancer group was considerably lower (37.4 ± 6.9 yr) than women in cancer group (51 ± 11.8 yr) (Table I).
Quantitation of HPV16 viral load in cervical pre-cancer and cancer tissues: To determine HPV16 viral load in cervical lesions, first HPV16URR real-time PCR was validated using 10- fold serial dilutions of WHO HPV16 International Standards diluted from stock in a dilution series of 50,000 to 5 Genome Equivalent per reaction in the background of about 50 ng HPV negative C33a DNA (Fig. 1). The technique was performed five times to determine the linearity of amplification. The assays were considered valid if correlation coefficient in each run was (r) ≥ 0.95. Specificity of desired amplification was confirmed using melt curve analysis. Although the assay could detect five copies per reaction at some instances, it could effectively and consistently detected 50 copies per reaction in a background of 50 ng of host DNA which is equivalent to ~7500 copies of diploid human genome. The HPV16 URR PCR was specific for HPV16 as no cross-reactivity was observed for sample previously genotyped as phylogenetically-related HPV31, HPV33, HPV52, HPV18 or HPV56 (data not shown).
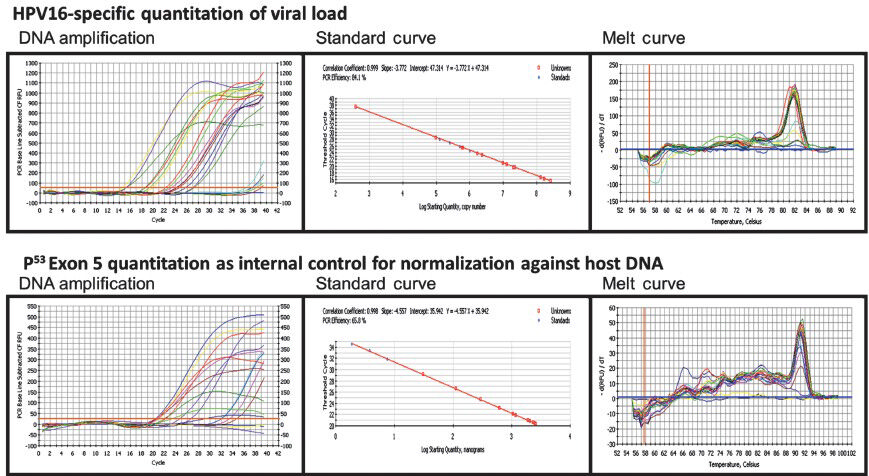
- Quantitative real-time PCR of HPV16 URR to determine HPV16 viral load in DNA isolated from cervical precancer and cancer tissue biopsies. Genomic DNA of HPV16 positive cervical precancer and cancer samples was amplified by type-specific HPV16 URR primers using Biorad iQ SYBR Green supermix (as described in Methods). Two-fold serial dilution of WHO HPV16 international standards starting from 5×104 copies/reaction in C33a DNA diluents were used as reference (upper panels). Amplification of p53 exon5 which was used as normalization control for genomic DNA input (lower panels). DNA amplification threshold cycle analysis (left panels); standard curve analysis for determining the efficiency of reaction and calculation of viral copy number and quantitation of host genome equivalents (middle panel); Melt curve analysis showing specificity of HPV16 and p53 exon 5 amplicons were performed in each run (right panel).
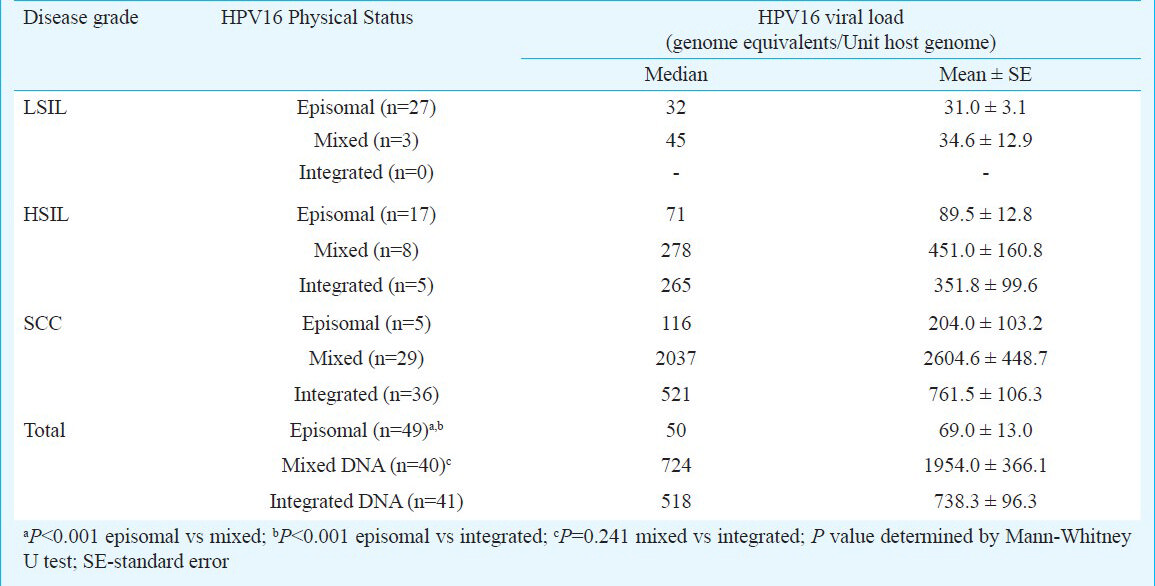
After validation, genomic DNA from 130 HPV16-positive cervical biopsies was analysed by real-time quantitative HPV16 URR PCR. Gross viral load thus obtained was normalized to input host DNA using p53 exon 5 amplification and the viral load was expressed as HPV16 genome equivalents per diploid host cell genome. Table II shows median, mean, interquartile and range of normalized HPV16 viral load in different tumour tissue types. The viral copy number in low-grade lesions ranged from 4 to 60 copies/cell equivalent (LSIL - median 33; interquartile 31) whereas in high-grade lesions it ranged between 29-1282 copies/cell equivalent (HSIL - median 116; interquartile 197). Analysis of viral loads between LSIL and HSILs revealed significant differences within precancer disease group (P< 0.001). On the other hand, tumour tissues of invasive cervical cancer lesions revealed a wide variation of HPV16 viral load that ranged from 68-7202 copies/cell equivalent (SCC - median, 628.50; interquartile 1651). These values were comparatively higher in majority of cancer cases than the viral load estimated in (P< 0.001). The distribution of viral load in cervical lesions of different histopathological grades revealed no grade-specific difference in HPV viral load except that HPV16 copy number was marginally higher in moderately differentiated MDSCC tissues (data not shown).
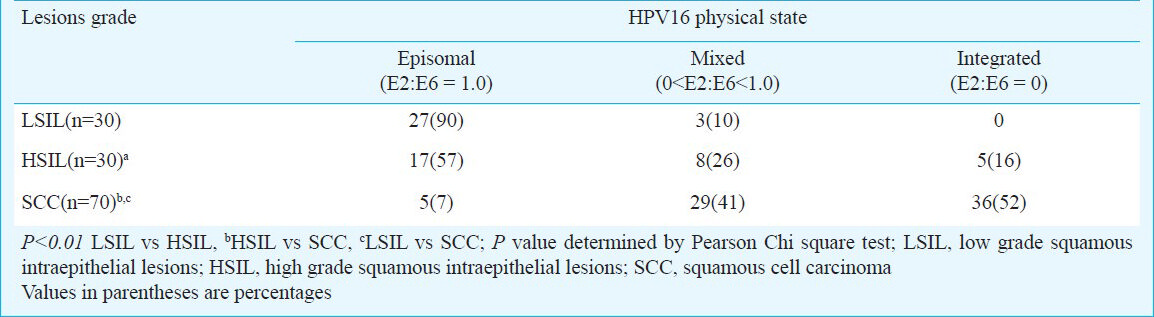
Determination of physical state of viral genome in HPV16 positive cervical precancer and cancer lesions: Physical state of HPV DNA in host genome was determined by comparing the specific and pre-validated PCR amplification of full length HPV16 E2 and HPV16 E6 regions. Mixes of vector-free HPV16 plasmid and SiHa cell DNA represented ratios from 1 to 0, were subjected to E2 and E6 PCR and E2:E6 ratios were calculated by densitometric analysis. A good correlation was observed between theoretical and experimental E2:E6 ratios when overall viral load was less than 50,000 genomes per reaction. At higher viral loads, the variability of the assay was higher when the E2: E6 ratios were more than 0.8. For assessment of genomic DNA of clinical cases, the E2:E6 ratios were further normalized to E2:E6 ratio of vector-free HPV16 plasmid DNA that was used in each dataset to control variations in PCR efficiencies (Fig. 2). Analysis of normalized E2:E6 densitometric ratios of HPV16-positive specimens demonstrated presence of predominantly episomal viral DNA in 90 per cent (27/30) of LSIL (Table III). Though none of these samples demonstrated fully integrated HPV16 genome, a small proportion (10%; 3/30) showed E2:E6 ratio less than 0.8 and were indicative of a mixed or concomitant form of the viral DNA. In contrast, 43 per cent of HSIL lesions had either fully integrated (16%; 5/30) or mixed form (26%; 8/30) of HPV16 DNA whereas 57 per cent (17/30) lesions has episomal HPV16 (Table III). Except one, all the HSIL cases with mixed HPV16 had E2:E6 ratio less than 0.8. On the other hand, 93 per cent of invasive cancers harboured either fully integrated (52%; 36/70) or mixed form (41%; 29/70) of HPV16 DNA. In a small proportion of cases, HPV16 was found in episomal state (7%; 5/70) and one case with mixed HPV16 was detected in borderline limits (E2:E6 ratio > 0.8).
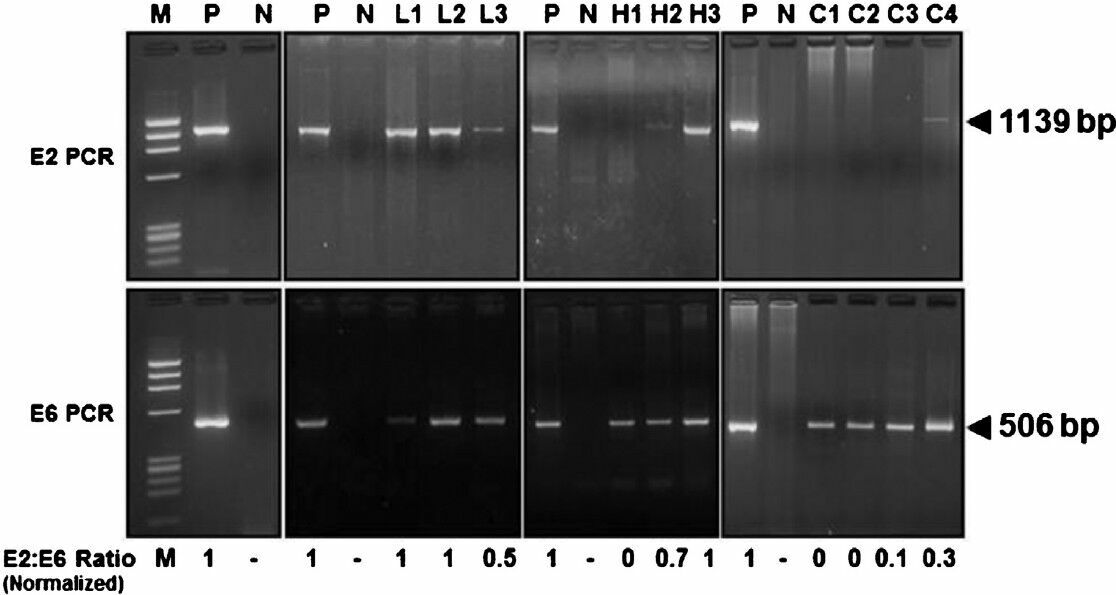
- Determination of physical state HPV16 genome in cervical precancer and cancer tissue biopsies by HPV16 E2 and HPV16 E6 amplification. HPV16 E2 and E6 open reading frame (ORFs) were amplified using specific primers, and E2:E6 densitometric ratio was determined as described in ‘Methods’. Vector-free HPV16 full length plasmid harbouring intact E2 and E6 region was used as positive control and as a reference for normalization of E2:E6 ratio in clinical samples. Upper panels showing gel photograph of HPV16 E2 amplification (1139bp) and lower panels show HPV16 E6 amplification (506bp) in DNA of vector control and clinical samples. Normalized E2:E6 ratio of the samples is indicated at the bottom of the respective lane. Normalized E2:E6 ratio = 0 represents completely integrated HPV16 genome, value = 1 represent episomal viral genome, and values between 0 and 1 indicate mixed form of HPV16 genome. M, φX174 HaeIII-digested molecular weight marker; P, HPV16 plasmid; N, PCR negative control (C33a genomic DNA); L1-L3, LSIL cases; H1- H3, HSIL cases; C1- C4, invasive cancer cases.
Association between HPV16 viral load and physical state of the virus: To assess whether varied levels of HPV16 viral load in different stage of cervical carcinogenesis have any ‘impact on’ or ‘association with’ the physical state of the virus in monotypic HPV16 infection, HPV16 viral load was analysed in cervical lesions harbouring HPV16 genome in different physical states. As depicted in Fig. 3, the copy number of HPV16 genome was comparatively higher in those tumour tissues where HPV16 was present in the mixed or in integrated state as compared to the viral load in tissues with HPV16 in episomal state. Interestingly, this trend was not specific to the lesion type (Table IV). When viral load in episomal state was evaluated among these tissue types, a marginal increase was observed in episomal viral load in HSIL (Median- 71, cases 17/30) and SCC lesions (Median- 116, cases 5/70) as compared to episomal viral load in LSIL tissues (Median- 32, cases 27/30) (Table IV). However, viral load in lesions harbouring purely episomal state were consistently lower as compared to precancer or cancer lesions with either mixed or integrated viral genome. Fully integrated HPV16 was not detected in any of the LSIL tissues examined. Interestingly, in SCC lesions with mixed state of HPV16 genome, the viral copies were the highest among all tissue types that harbour either fully integrated or predominantly episomal state of HPV16 genome (Median viral copies of mixed state- (2037) vs. integrated state- (521) (GE/unit host genome). SCC lesions when further stratified with respect to their histopathological grades to assess association of HPV16 physical state with the viral load, these lesions did not show much difference on these parameters (data not shown).
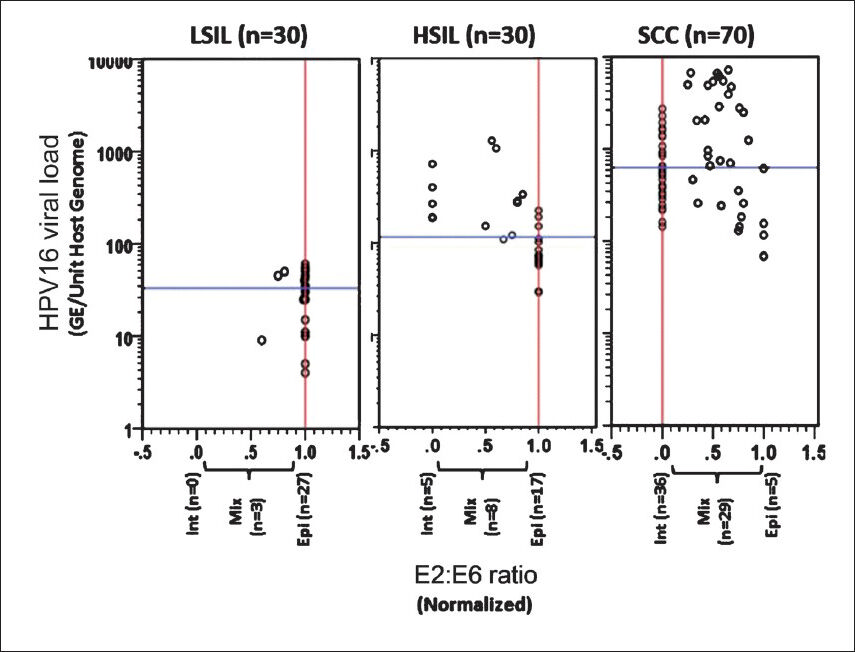
- Association of HPV16 physical state with the viral load in different precancer and cancer cases. Distribution of normalized HPV16 E2:E6 ratio and HPV 16 viral load in cervical precancer (LSIL and HSIL) and cancer cases. Each circle indicates individual case. Normalized E2:E6 ratios and viral load were calculated as described in Methods. Normalized E2:E6 = 0 represents completely integrated HPV16; E2:E6 = 1 represents episomal viral genome, E2:E6 between 0 and 1 indicates concomitant HPV16 genome. Vertical red line and horizontal blue line represent median values of E2:E6 ratio and HPV16 viral load in each disease group, respectively.
Discussion
HPV viral load and physical state of the viral genome are important determinants of HPV infection which influence the tumorigenic transformation of normal cervical epithelium and progression of the disease11, however, variability in results of various studies conducted on these parameters have limited their clinical use. Among others, infection with multiple HPV genotypes is the major confounding variable. Therefore, in the present study, we examined copy number and physical state of infecting monotypic high risk papillomavirus type 16 in cervical precancer and cancer lesions. Analysis of the viral load in these lesions revealed occurrence of a quantitative increase in viral genomes with disease severity. HPV16 viral load was found to be highest in cancer cases. Our full length E2 PCR amplification-based approach revealed a distinct distribution of physical state of HPV16 genome in samples representative of three different stages of natural history of cervical carcinogenesis which was suggestive of integration event as early as in LSIL (10%) that accumulated during progression of the lesions to HSIL (42%) and reached the highest in invasive cancers (93%). Despite the integration event, in a major proportion of the precancer and cancer tissues HPV16 genome was apparently detected in concomitant or mixed state where both integrated and episomal form of HPV16 genome co-existed.
Our investigation demonstrated a gradual increase in HPV16 viral load in progressive cervical lesions, i.e. LSIL to HSIL and pre-cancer to cancer. Cancer lesions showed the highest viral load. The association between the viral load and the different stages of cervical disease is still debated. An elevated HPV16 viral load is associated specifically with high-grade cervical lesions152122, and a 43-fold increased risk of cancer has been reported in cases with high viral load7. However, a viral load higher in LSIL than in HSIL and lower in HSIL than that in carcinoma14 as well as no association between HPV16 viral load and disease progression2324 have also been reported. Studies using a wide range of study designs, laboratory assays, and analytical methods have suggested that quantitative measurement of viral loads, especially for the highly oncogenic HPV16, may be associated with high grade lesions and disease progression4711141722232526. Most of the early analyses of viral load did not adjust for specimen cellularity, number of atypical cells, multiple infections with different HPV types. Therefore, these studies varied with respect to the magnitude of the viral load in different types of cervical lesions. Majority of these issues have been appropriately taken care of in the present investigation by using (i) internal control with similar size of amplicon to minimize PCR efficiency issues, (ii) use of HPV16 International Standards with known copy number as calibrator, (iii) use of quantitative real-time PCR for calculation of absolute copy number of viral DNA and (iv) a uniform reference to host cell diploid DNA content which made the analysis more robust and reliable as it could overcome possible sources of variation. However, in precancer cases, where the proportions of tissue specimen may consist of normal potentially uninfected cells, the viral loads measured may be an underestimation for HPV16-infected cells.
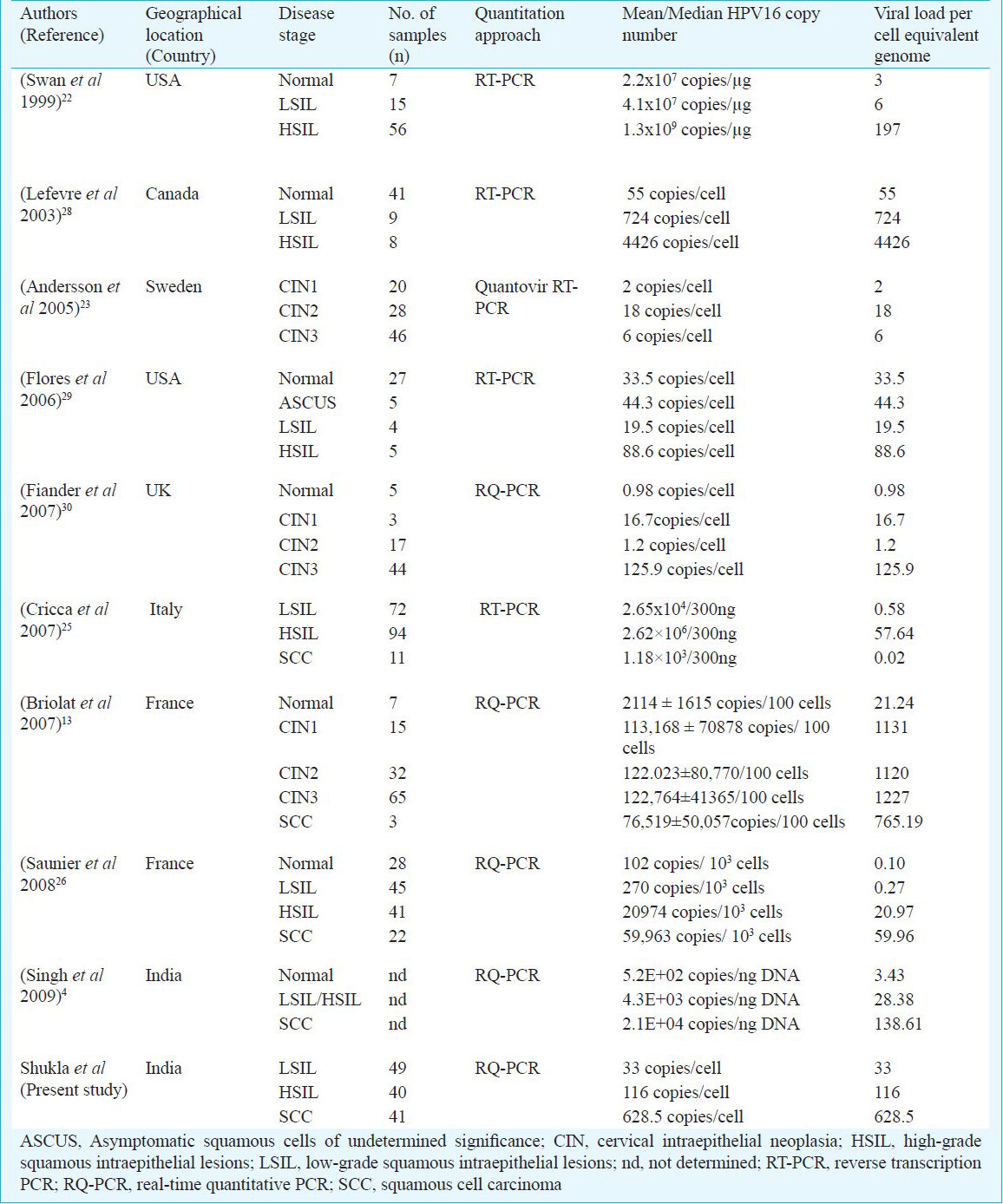
Early studies, whether semi-quantitative27 or quantitative1322232526282930, demonstrated a correlation of HPV viral load with disease progression. Majority of these studies had strong inter-study variations in disease stage-specific viral loads which may partly be related to geographical HPV16 variations (Table V). In some studies, however, HPV16 viral load ranges and trends resembled to that reported in the present study4132730, however, the load detected in the present study could not be quantitatively compared with these studies as we did not analyze HPV16 loads from non-Indian women. Moreover, the direct comparison of the viral load values among various studies would not be an ideal approach to explain the differences in view of the selective nature of sample inclusion (selection of only HPV16 infected samples) of the present study. Our observations indicate a strong association of type-specific HPV16 viral load with disease severity provided the confounding variables like co-infection with other HR-HPVs are avoided.
Apart from plurality of infecting HR-HPV types which may have different biological behaviour in the host cell, age of patient harbouring these infections may also contribute significantly to variations in viral load. Women older than 30 yr showed significantly higher viral loads in HSIL as compared to LSILs for both HPV16 and HPV1821. However, such correlations were not seen in women infected with these HR-HPVs who were below 30 yr of age and were reported to have transient infections31. HR-HPV infections in women beyond 30 yr are considered to have higher tendency for persistence32 and this may be responsible for integration of viral DNA in host genome which provides stability to the infection. Therefore, gradual increase in viral copies in progressive lesions could be playing a role in determining the integration of virus in women aged 30 yr. As majority of the cases enrolled in our study were of age above 30 yr, the age of the patients could be the most probable reason of finding a strong correlation of HPV16 viral load with increasing severity of cervical lesions. However, among the study subject, a significant difference was observed in mean age of patients of precancer and cancer disease group. As we did not have any corresponding age-matched specimen in precancer vs. cancer group, the effect of age on the study parameters could not be assessed and the age difference could not be accounted for the analysis.
In the present study, LSILs were found to harbour predominantly the episomal HPV16, the proportion of which gradually declined in high grade lesions with increasing disease severity. Occurrence of episomal HPV16 in LSILs has been demonstrated in several studies with highly variable degree15182325. Reasons for such a variation in episomal form of HPV16 during LSIL, which reflects an early stage of viral infection, are not known but could be attributed to variations in analysis as well as the patient population analyzed. Similarly, variations in frequency of episomal HPV16 in HSIL3334 and in cervical carcinoma where it ranged upto 45 per cent have been documented1226. Different HPV16 variants are known to express different integration frequencies24 and minor sequence variations in different geographical isolates may be in part responsible for variable frequencies of cancer cases found to harbour HPV in different physical forms.
The frequency of samples with integrated HPV16 genome in cancer cases in our study was slightly lower (52%) compared to our earlier report (69%)18. However, when cancer tissues harbouring the concomitant form of HPV16 which also consisted of integrants were collectively considered, majority (93%) of samples indicated occurrence of integration event. Therefore, low frequency integration events with deleted E2 in a background of episomal or integrated concatameric HPV16 cannot be ruled out in samples with predominantly episomal HPV16. These observations suggested that integration could be a common event in HPV16 in Indian population, but elimination of episomal forms may not be essential during tumorigenic transformation and there could be some selective advantage of the episomal forms in concomitant state for persistent and progressive HPV infection. Other studies from different regions of India also demonstrated pure integrant frequency of HPV16 in cervical cancer cases ranges from 32-38 per cent of cancer cases3536. Frequencies of finding purely integrated HPV16 genome in malignant cervical carcinomas is variable and range from as low as 30 per cent to as high as 100 per cent in different studies globally121517252633. Apart from technical issues, prevalent HPV16 variants could be the major factor that could possibly contribute to the preference of the virus to maintain a high copy number concomitant state24.
Presence of mixed form of HPV16 genome in a significant proportion of cervical carcinoma cases in the present study suggests that requirement of E2 disruption for induction of viral oncogenes may be a redundant event for HPV16-induced cervical carcinogenesis and indicative of an alternate mechanism(s) for overriding E2-mediated inhibitory effects. Association between HPV16 viral load and its physical state found in the present study demonstrated that the median copy number of HPV16 genome increased in those cases where its genome was present primarily in mixed or integrated form in comparison to episomal form. HPV16 copy number was marginally higher in lesions harbouring mixed form than the tissues with fully integrated HPV16 genome. Many reports demonstrated simultaneous analysis of viral load and physical state of HPV16 genome by analyzing E2 status14152526333437, but these reports mainly assessed importance of viral load and physical state in disease prognosis and none of these analyzed a correlation between these important elements of HPV16 infection and their potential impact on each other. Some of these studies have documented higher viral copy number in mixed form of HPV16 physical state in comparison to episomal and integrated form in SCC142534. The reason why cervical lesions with mixed state of HPV16 genome have high viral loads is not known.
Presence of intact E2 in cervical lesions in no way suggests its protein expression in the lesion. Recent evidences using HPV16 E6-specific antibodies have revealed a low or undetectable E2 in cervical cancer tissues3839. However, it is also important to note that presence of an intact E2 gene and frequent reporting of E2 transcripts in cervical lesions3840 indicate a longer E2 expressing phase of viral infection in lesions with intact E2 as compared to E2-disrupted HPV16 infections. This prolonged E2 expression might have not only increased the viral copy number but reduced the chances of spontaneous regression of the lesion during the course of disease progression. On the other hand, E2 expression in early precancer stages is well established. However, these lesions (LSIL and HSIL) were found harbouring low copy number of HPV16 genome despite the fact that most of these lesions were detected with intact E2. The low copy number may be ascribed to stronger cellular control over DNA replication or alternatively, the difference could be partly due to relatively lower percentage of infected cells in these precancer lesions that might have resulted in underestimation of viral copy number.
In conclusion, the present study demonstrated an increase in frequency of integration and the viral load during HPV16-mediated cervical carcinogenesis. Moreover, loss of E2 region of HPV16 genome or elimination of episomal forms of virus was not required in a major proportion of invasive cervical cancer cases. These leads will be useful in understanding the relation of viral load and integration events in disease progression and thus will help in designing of individualized therapeutic strategies to target viral replication and integration events.
References
- A high frequency of human papillomavirus DNA sequences in cervical carcinomas of Indian women as revealed by Southern blot hybridization and polymerase chain reaction. J Med Virol. 1992;36:239-45.
- [Google Scholar]
- Human papilloma virus genotyping, variants and viral load in tumors, squamous intraepithelial lesions, and controls in a north Indian population subset. Int J Gynecol Cancer. 2009;19:1642-8.
- [Google Scholar]
- Infection of human papillomaviruses in cancers of different human organ sites. Indian J Med Res. 2009;130:222-33.
- [Google Scholar]
- Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int J Cancer. 2003;106:396-403.
- [Google Scholar]
- Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2189-93.
- [Google Scholar]
- Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol. 2007;212:356-67.
- [Google Scholar]
- Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proc Natl Acad Sci USA. 2006;103:3822-7.
- [Google Scholar]
- Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet. 2002;360:228-9.
- [Google Scholar]
- The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11-22.
- [Google Scholar]
- Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307-13.
- [Google Scholar]
- HPV prevalence, viral load and physical state of HPV-16 in cervical smears of patients with different grades of CIN. Int J Cancer. 2007;121:2198-204.
- [Google Scholar]
- Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J Clin Pathol. 2006;59:513-7.
- [Google Scholar]
- The use of viral load as a surrogate marker in predicting disease progression for patients with early invasive cervical cancer with integrated human papillomavirus type 16. Am J Obstet Gynecol. 2009;201:79 e1-7.
- [Google Scholar]
- Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J Clin Microbiol. 2006;44:1755-62.
- [Google Scholar]
- Association of viral load with HPV16 positive cervical cancer pathogenesis: Causal relevance in isolates harboring intact viral E2 gene. Virology. 2010;402:197-202.
- [Google Scholar]
- Analysis by polymerase chain reaction of the physical state of human papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J Gen Virol. 1992;73:2327-36.
- [Google Scholar]
- World Health Organization. Human papillomavirus laboratory manual. (1st ed). Geneva: WHO; 2010. p. :35-63.
- [Google Scholar]
- Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402-8.
- [Google Scholar]
- Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J Med Virol. 2006;78:1131-40.
- [Google Scholar]
- Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol. 1999;37:1030-4.
- [Google Scholar]
- Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN) Br J Cancer. 2005;92:2195-200.
- [Google Scholar]
- Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706-12.
- [Google Scholar]
- Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol. 2007;106:549-57.
- [Google Scholar]
- Analysis of human papillomavirus type 16 (HPV16) DNA load and physical state for identification of HPV16-infected women with high-grade lesions or cervical carcinoma. J Clin Microbiol. 2008;46:3678-85.
- [Google Scholar]
- Use of semi-quantitative PCR for human papillomavirus DNA type 16 to identify women with high grade cervical disease in a population presenting with a mildly dyskaryotic smear report. Br J Cancer. 1993;67:602-5.
- [Google Scholar]
- Real-time PCR assays using internal controls for quantitation of HPV-16 and beta-globin DNA in cervicovaginal lavages. J Virol Methods. 2003;114:135-44.
- [Google Scholar]
- Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118:1187-93.
- [Google Scholar]
- Variation in human papillomavirus type-16 viral load within different histological grades of cervical neoplasia. J Med Virol. 2007;79:1366-9.
- [Google Scholar]
- Human papillomavirus infection is transient in young women: a population-based cohort study. J Infect Dis. 1995;171:1026-30.
- [Google Scholar]
- A prospective study of age trends in cervical human papillomavirus acquisition and persistence in Guanacaste, Costa Rica. J Infect Dis. 2005;191:1808-16.
- [Google Scholar]
- Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886-91.
- [Google Scholar]
- Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol. 2004;92:873-80.
- [Google Scholar]
- HPV16 E2 gene disruption and polymorphisms of E2 and LCR: some significant associations with cervical cancer in Indian women. Gynecol Oncol. 2006;100:372-8.
- [Google Scholar]
- Human papillomavirus 16 E6/E7 transcript and E2 gene status in patients with cervical neoplasia. Mol Diagn. 2004;8:57-64.
- [Google Scholar]
- Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res. 2009;69:3828-32.
- [Google Scholar]
- Loss of HPV16 E2 protein expression without disruption of the E2 ORF correlates with carcinogenic progression. Open Virol J. 2012;6:163-72.
- [Google Scholar]
- HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 2010;70:5316-25.
- [Google Scholar]
- Diagnosing cervical cancer and high-grade precursors by HPV16 transcription patterns. Cancer Res. 2010;70:249-56.
- [Google Scholar]






