Translate this page into:
Neonatal septicaemia caused by diverse clones of Klebsiella pneumoniae & Escherichia coli harbouring blaCTX-M-15
Reprint requests: Dr Sulagna Basu, Division of Bacteriology, National Institute of Cholera & Enteric Diseases (ICMR), P33, CIT Road, Scheme XM, Beliaghata, Kolkata 700 010, India e-mail: supabasu@yahoo.co.in
-
Received: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background & objectives:
Information about the genetic diversity of the extended-spectrum β-lactamases (ESBLs) and the clonal relationship of the organisms causing neonatal infections is limited, particularly from India where neonatal mortality is high. This study was undertaken to investigate the molecular epidemiology and risk factors associated with neonatal septicaemia caused by ESBL-producing Klebsiella pneumoniae and Escherichia coli.
Methods:
Bloodstream isolates (n=26) of K. pneumoniae (n=10) and E. coli (n=16) from the neonates admitted in a tertiary care hospital in New Delhi during January to May 2008 were characterized. Antimicrobial susceptibility tests were carried out and ESBL production was assessed phenotypically. PCR was carried out for ESBL and ampC genes. Genotyping was performed by pulsed-field gel electrophoresis (PFGE). Conjugation experiments were done to determine the mobility of ESBL genes. Risk factors associated with ESBL-producing K. pneumoniae and E. coli infections were analysed.
Results:
Resistance rates to most of the antibiotics tested were high, except for imipenem. Among the isolates tested, 60 per cent of K. pneumoniae and 75 per cent of E. coli were ESBL producers. PFGE of the isolates demonstrated a vast diversity of genotypes with no epidemic clones. Despite the clonal diversity, blaCTX-M-15 was detected in 100 per cent of ESBL-positive isolates. The other genes present in ESBL-positive isolates were blaTEM-1, blaSHV-1, blaSHV-28, blaSHV-11, and blaSHV-12. Class 1 integrons were detected in 7 of 18 ESBL-positive isolates. Moreover, the plasmid carrying blaCTX-M-15, in E. coli and K. pneumoniae were self transferable. Feeding through an enteral tube was identified as the only risk factor for sepsis by ESBL-producing organisms.
Interpretation & conclusions:
The study emphasises the presence of blaCTX-M-15 in clonally diverse isolates indicating probable horizontal transfer of this gene. The widespread dissemination of CTX-M-15 is of great concern as it further confines the limited therapeutic interventions available for neonates.
Keywords
CTX-M-15
diverse clones
ESBLs
Escherichia coli
Klebsiella pneumoniae
neonatal sepsis
risk factor
Neonatal mortality in developing countries like India is very high12. Microbial infections leading to sepsis is a major contributor to neonatal deaths in the developing world3. The overall fatality rate due to neonatal sepsis and pneumonia in developing countries is estimated to be about 25 per cent, based largely on data for infants treated in hospitals3. Developing countries like India, Nigeria, Democratic Republic of the Congo, Pakistan, and China were responsible for large proportions of the global total for neonatal death due to sepsis4.
An important compounding factor in the treatment of these infections associated with sepsis is the emergence of extended-spectrum beta lactamase (ESBL)-producing Enterobacteriaceae5. ESBLs are derived from genes by mutation in the narrower-spectrum TEM-1, TEM-2 or SHV-1 β-lactamases67. Additionally, other types of ESBLs (CTX-M) that are plasmid-mediated, exhibit an overall preference for cefotaxime and ceftriaxone8. At present, more than 100 genetically distinct TEM, SHV, OXA and CTX-M-type ESBLs have been characterized (http://www.lahey.org/studies/webt.asp).
Among Gram-negative bacilli the two most important bacterial pathogens causing neonatal sepsis in developing countries are Klebsiella pneumoniae and Escherichia coli910. Studies on ESBL-mediated resistance mechanisms in adult patients from developing countries have been carried out1112131415161718. However, studies on the types of ESBLs and the clonal relationship of the organisms producing them, especially in neonatal infections from India are limited192021.
In this study, isolates of K. pneumoniae and E. coli causing bloodstream infection from neonates admitted to a tertiary care centre in New Delhi, India were analysed. The aim of the study was to carry out (i) molecular characterization of the ESBL genes in E. coli and K. pneumoniae isolates involved in neonatal sepsis, (ii) assessment of any significant differences in the clinical or demographical variables in the neonates that predispose them to acquisition of ESBL-producing organisms, (iii) determination of the molecular typing of the ESBL-producing isolates to ascertain the clonal carriage of the ESBL genes.
Material & Methods
Clinical setting and bacterial isolates: Isolates included in the study were collected from septicaemic neonates during January to May 2008 from Department of Microbiology at Vardhman Mahavir Medical College and Safdarjang Hospital, tertiary care centre in New Delhi. This centre has two nurseries, one for intramural admissions (born at the hospital) and one for extramural admissions (born outside the hospital). During the study period, a total of 1443 neonates were admitted to the nurseries, among whom, 405 neonates were suspected to have sepsis. Of these 405, 177 (43.7%) neonates were culture positive. Among these 177 neonates, 63 (35.59%) had aerobic Gram-negative bacilli (GNB) in their blood. Of the 63 GNBs, K. pneumoniae (n=16) and E. coli (n=10) were isolated. All K. pneumoniae and E. coli isolates obtained during this period were included in the study. Details of the neonates from whom K. pneumoniae and E. coli have been isolated are included in Table I.
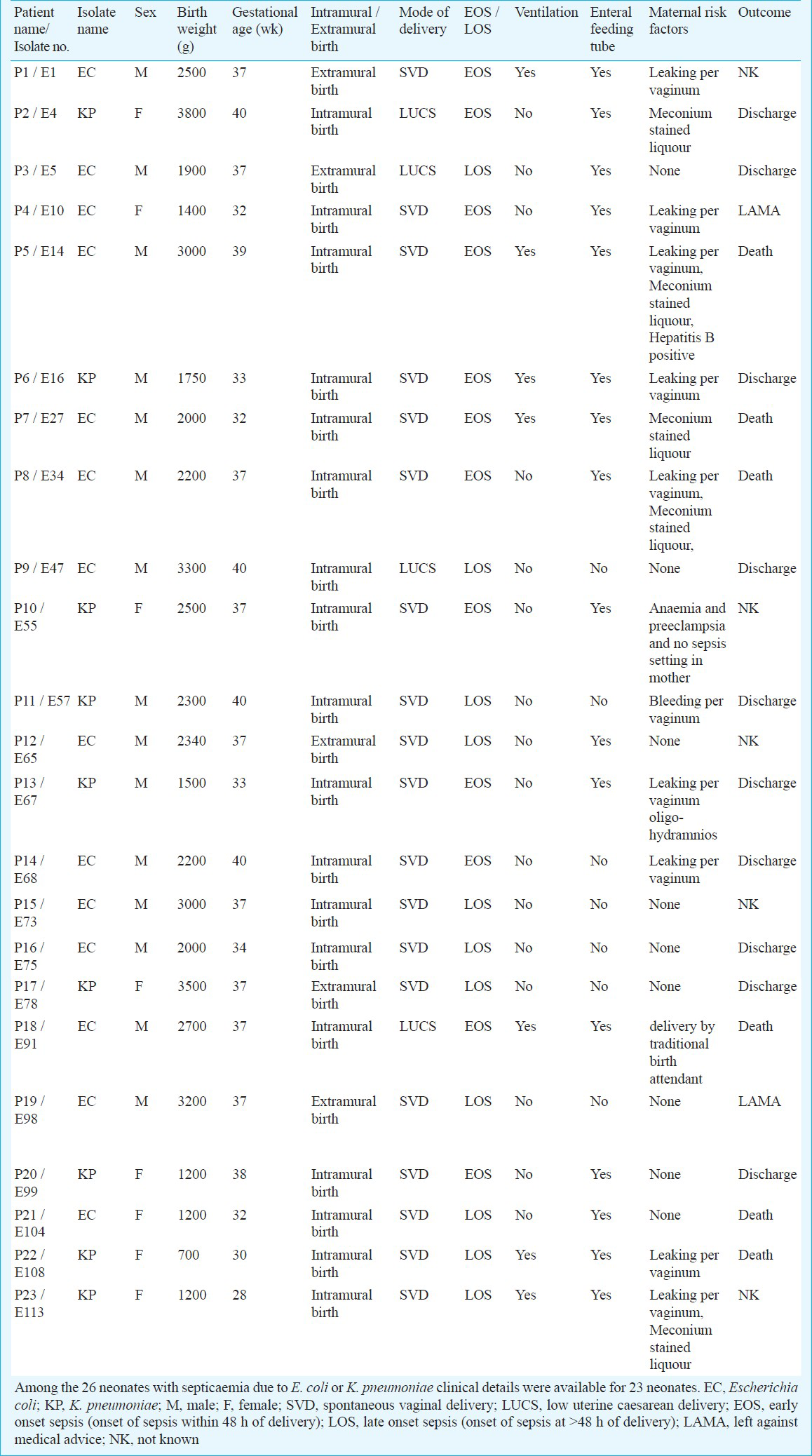
Blood specimen collection and identification of the isolates: One ml blood for culture was drawn with aseptic precautions from a peripheral vein. Cultures were processed in BacT/ALERT 3D system (bioMeérieux, Marcy l’Etoile, France). For any culture which flagged positive, Gram stain was performed and subculture was done on appropriate medium based on the Gram stain: MacConkey agar and 5 per cent sheep blood agar for Gram-negative and Gram-positive organisms, respectively. Bottles were incubated in the system for up to seven days, at the end of which all negative bottles were subcultured once on blood agar before discarding. The isolates were identified by different biochemical tests and further verified by using an ID32E or ID32GN system (bioMeérieux).
Antibiotic susceptibility and MIC determination: Antibiotic susceptibility was determined as per Clinical and Laboratory Standards Institute (CLSI) criteria22. E. coli ATCC 25922 was used as quality control strain. The minimum inhibitory concentration (MIC) values (μg/ml) for cefotaxime, ceftazidime and cefepime were determined using E-tests (AB Biodisk, Solna, Sweden) and interpreted according to manufacturer's instructions.
ESBL confirmatory test: ESBL production was confirmed using the cephalosporin/clavulanic acid (cefotaxime/clavulanic acid and ceftazidime/clavulanic acid) combination disk test22. Fifty per cent of the isolates that were positive in the combination disk test were again confirmed by the double disk synergy test5.
PCR detection and sequencing of β-lactamases & integrons: Genotypic analysis for blaSHV, blaTEM, blaOXA-1 and blaCTX-M was performed for isolates found to be positive in ESBL confirmatory test and subsequently whole open reading frame (ORF) of PCR products were amplified for sequencing232425. PCR controls for blaSHV and blaTEM were K. pneumoniae ATCC 700603 and E. coli ATCC 35218, respectively. Control DNAs were used for detection of three groups of blaCTX-M genes (provided by Dr Guillaume Arlet, Service de Bacteriology, Hospital Tenon, France). PCR was carried out to detect class 1, 2 and 3 integrons in all isolates and presence of plasmid-mediated ampC genes was also checked in cefoxitin-resistant isolates using PCR2627. PCR controls were used for ampC genes (MIR-1, DHA-1, ACT-1 and FOX-5) (kind gift of George A. Jacoby, Lahey clinic, Massachusetts). PCR products were purified using QIAquick PCR Purification Kit (Qiagen, USA), sequenced directly on both strands using the BigDye Terminator v3.1 Cycle Sequencing Kit and analysed with an automated sequencer ABI3100 genetic analyser (Applied Biosystems, Foster City, CA). Sequence analysis was performed using the Lasergene DNASTAR sequence analysis software (DNAStar, Madison, USA). The BLASTN program was used for database searching (http://www.ncbi.nlm.nih.gov/BLAST/).
Pulsed-field gel electrophoresis (PFGE): PFGE was performed following the Pulse Net protocol (www.cdc.gov/pulsenet/protocols.htm). Overnight cultures of the isolates on Tryptic Soy Agar with 5 per cent sheep blood were suspended in the cell suspension buffer (100mM Tris, pH8.0, 100mM EDTA, pH 8.0). The optical density (600 nm) was adjusted to 1.3-1.4. Plugs were prepared by mixing 200 μl of the bacterial suspension [with 10 μl of Proteinase K (Takara, Japan)] with 200 μl of 1 per cent plug agarose (Seakem Gold Agarose, Cambrex, USA). Cell lysis was carried out in lysis buffer (50mM Tris: 50mM EDTA, pH 8.0, 1% Sarcosyl, 100 μg/ml Proteinase K) at 50°C. Digestion of DNA was performed by overnight incubation of the plug in the presence of 50 U/plug of the restriction endonuclease Xba I (New England Biolabs, USA) at 37°C. Electrophoresis was performed in a 1 per cent agarose (Pulse certified agarose, BioRad, Hercules, CA, USA) on a CHEF DRIII apparatus (Bio-Rad Laboratories) with 6V/cm for 19 h at 14°C with an initial switch time of 2.2 to 52.0 sec. Salmonella Braenderup H9812 was included as a molecular weight marker. XbaI macrorestriction patterns were compared visually and interpreted according to the criteria of Tenover et al28.
Conjugation: Conjugal transfer of blaCTX-M to sodium azide-resistant E. coli J53 recipient (provided by George A. Jacoby, Lahey clinic, Burlington, Massachusetts, USA) was attempted by a broth mating assay29. Transconjugants were selected on Luria-Bertani agar plates containing cefotaxime (4 μg/ml) and sodium azide (100 μg/ml). The approximate sizes of the plasmids in the test isolates and transconjugants were estimated using logarithmic plots, generated with plasmids of known molecular mass30. E. coli K12 V517 (sizes of the plasmids are 54.2, 7.2, 5.6, 5.1, 2.7 & 2 kb) and Shigella flexneri YSH 6000 (sizes of the plasmids are 212, 3.9 & 2.7 kb) were used as reference standards. The presence of blaCTX-M in the transconjugants was confirmed by PCR.
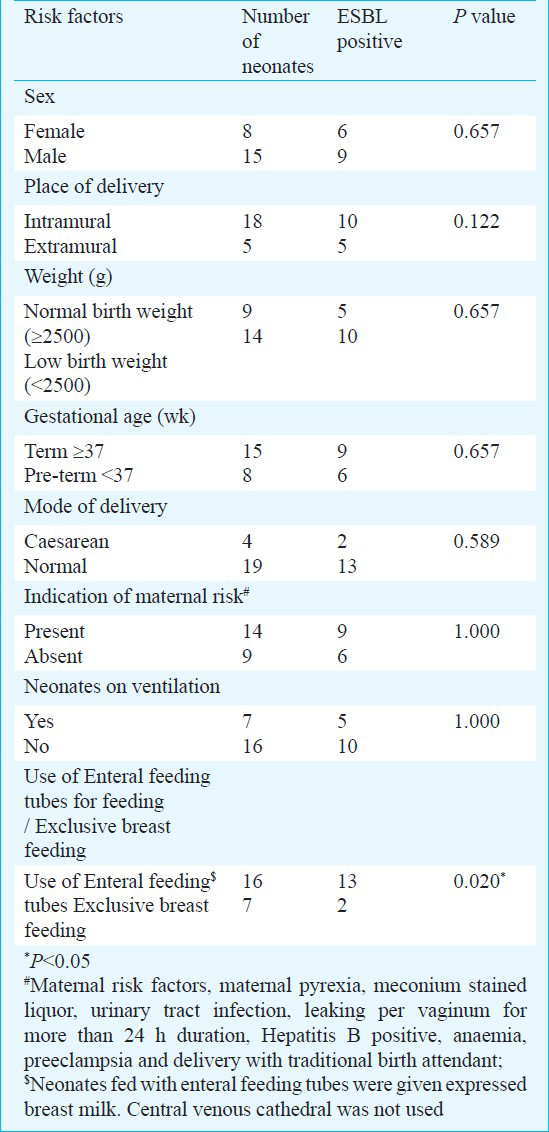
Statistical analysis: Fisher's Exact test was used for the assessment of risk factor for sepsis by comparing variables. This included sex, gestational age, birth weight, place of delivery (intramural/extramural), mode of delivery, use of a mechanical ventilator, type of feeding (feeding of expressed breast milk by enteral feeding tubes or breast feeding) and maternal risk factors (maternal pyrexia, meconium stained liquor, urinary tract infection, leaking per vaginum for more than 24 h duration, Hepatitis B positive, anaemia, preeclampsia and delivery of baby by traditional birth attendant). All comparisons were unpaired and all tests of significance were two-tailed.
Results
In comparison to K. pneumoniae, isolates of E. coli0 demonstrated higher resistance to the β-lactamase stable β-lactams, e.g. 3rd generation cephalosporins (cefotaxime, ceftazidime, cefpodoxime, and ceftriaxone), monobactams (aztreonam), aminoglycoside (gentamicin), and fluoroquinolone (ciprofloxacin). On the other hand, K. pneumoniae isolates showed higher resistance than E. coli against ampicillin, piperacillin, amikacin and netilmicin. Only imipenem exhibited 100 per cent activity in both isolates. Rates of resistance to β-lactam and non-β-lactam antibiotics were as follows (in K. pneumoniae versus in E. coli, respectively): ampicillin, 100 vs 88 per cent; piperacillin, 100 vs 88 per cent; cefotaxime, 60 vs 75 per cent; ceftazidime, 30 vs 50 per cent; cefpodoxime, 60 vs 75 per cent; ceftriaxone, 60 vs 75 per cent; azteonam, 60 vs 75 per cent; amikacin, 40 vs 25 per cent; gentamicin, 60 per cent vs 75 per cent; netilmicin, 50 vs 19 per cent; and ciprofloxacin, 60 vs 91 per cent. Cefoxitin resistance was low in both K. pneumoniae [30% (3/10)] and E. coli [6% (1/16)] isolates. All ESBL-negative isolates showed MIC values for cefotaxime, ceftazidime and cefepime in the range of 0.094 - 0.5 μg/ml and values for ESBL-positive isolates are presented in Table II.
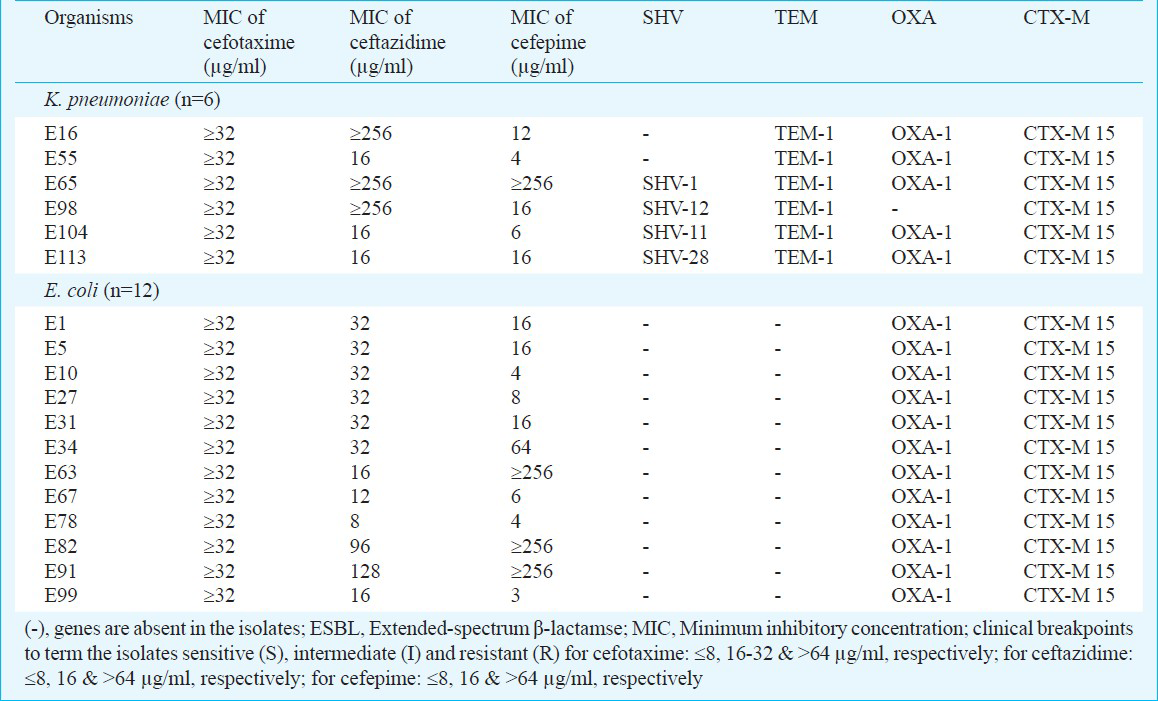
The overall percentage of ESBL-positive isolates in cephalosporin/clavulanic acid disk test was 60 per cent (6/10) for K. pneumoniae and 75 per cent (12/16) for E. coli. Isolates of K. pneumoniae and E. coli which were positive for ESBL by phenotypic tests showed presence of blaCTX-M group 1. Among ESBL-positive E. coli (n=12), blaOXA-1 was present in 100 per cent of isolates but none showed the presence of blaSHV and blaTEM. ESBL-positive K. pneumoniae (n=6) isolates possessed blaOXA-1[83% (5/6)], blaTEM [100% (6/6)] and blaSHV [67% (4/6)]. Sequencing revealed the presence of blaTEM-1, blaSHV-1, blaSHV-28, blaSHV-11, blaSHV-12 and blaCTX-M-15 in ESBL-positive isolates (Table II). Plasmid- mediated ampC β-lactamases could not be detected in any of the cefoxitin-resistant isolates (n=4).
Only seven ESBL-positive isolates (E5, E31, E55, E78, E98, E104 & E113) were found to possess class 1 integron which was confirmed by sequencing. However other classes of integrons could not be identified in any of these strains. Molecular typing of K. pneumoniae (n=10) revealed diversity (Fig. 1). Among E. coli (n=16) isolates, one clonal cluster was identified which consisted of three isolates (E5, E10 & E31). Another cluster of E. coli consisted of two (E27 & E34) closely related isolates (Fig. 2). CTX-M-15 was detected in all isolates that were ESBL-positive irrespective of their genotypes.
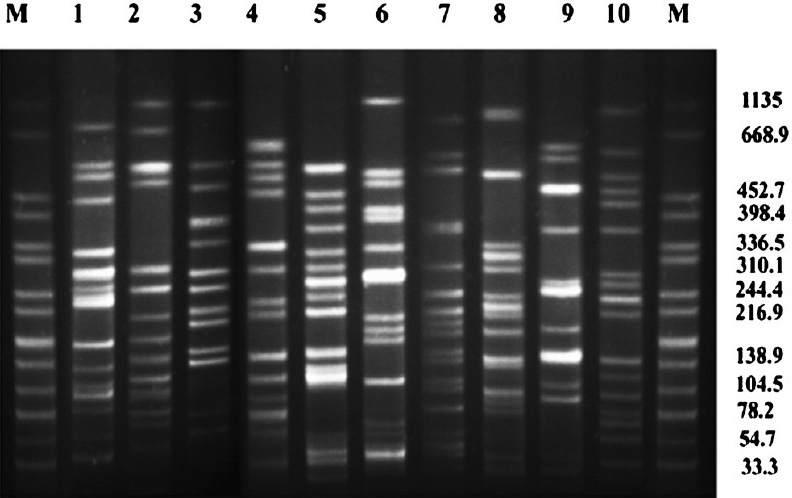
- Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA of CTX-M-15 producing K. pneumoniae (Lane 1-10) isolated from blood of neonates. Lane M: Salmonella serotype Braenderup H9812 as reference standard (band sizes in kilobases).
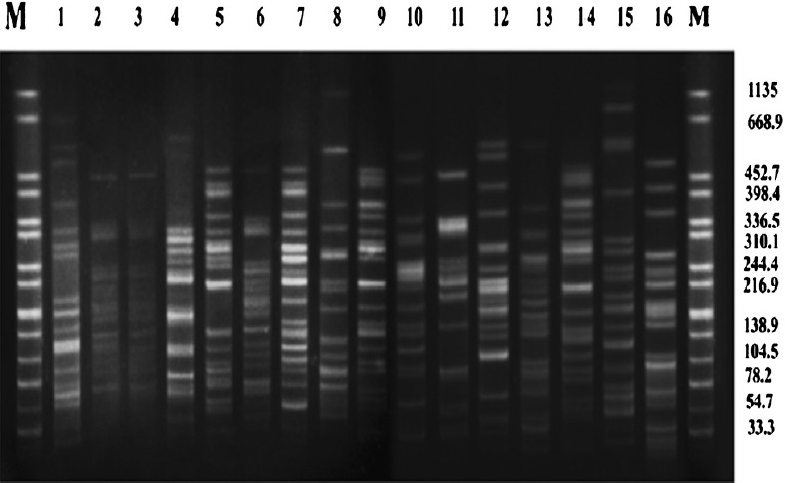
- Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA of CTX-M-15 producing E. coli (Lane 1-16) isolated from blood of neonates. Lane M: Salmonella serotype Braenderup H9812 as reference standard (band sizes in kilobases).
Different sizes of megaplasmids (~54.2 Kb upto >212 Kb, data not shown) were isolated from ESBL-positive isolates (12 E. coli & 6 K. pneumoniae) hourbouring blaCTX-M-15 gene. Conjugal transfer of blaCTX-M-15 gene carried out for one isolate each of E. coli and K. pneumoniae was successful. The presence of blaCTX-M-15 was confirmed by PCR in the transconjugants and plasmid analysis indicated that these had acquired megaplasmids present in the donor isolates (Fig. 3).
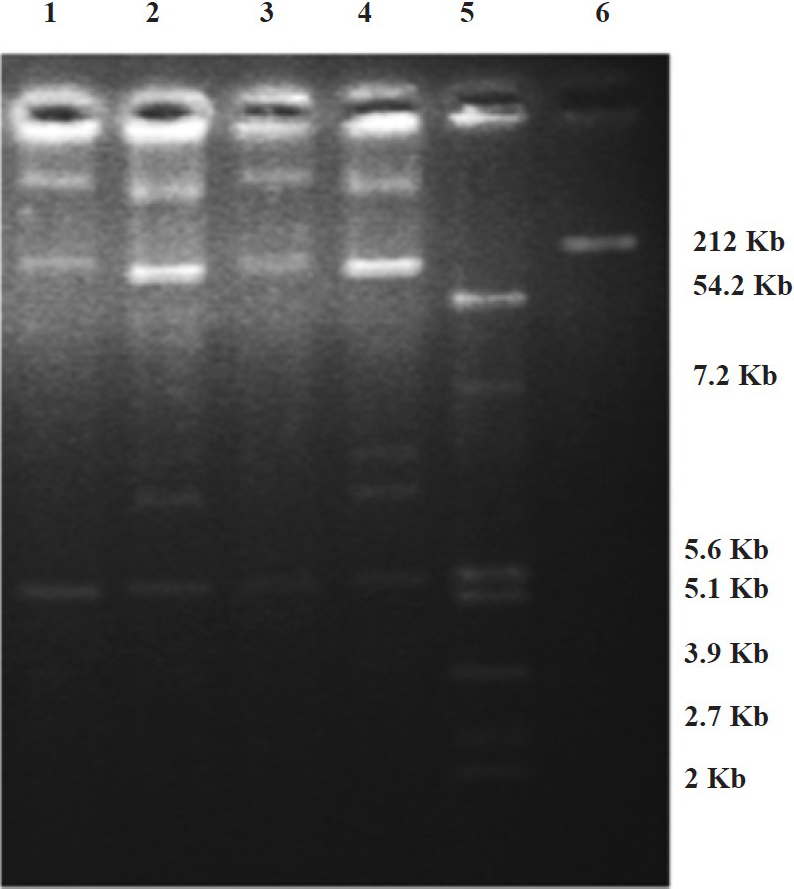
- Plasmid profiles of test isolates and transconjugants of E. coli and K. pneumoniae hourbouring blaCTX-M-15. Lanes 1 & 2: transconjugant of E. coli and test isolate of E. coli from blood of neonates, respectively. Lanes 3 & 4: transconjugant of K. pneumoniae and test isolate of K. pneumoniae from blood of neonates, respectively. Lanes 5 & 6: E. coli K12 V517 and Shigella flexneri YSH 6000 were used as reference standards respectively.
Risk factors for sepsis due to ESBL-producing organisms were examined for neonates (Table III). Among all the factors analysed for acquisition of ESBL-producing organisms, use of enteral feeding tubes was found to be the only significant risk factor (P=0.02). No significant association was found between ESBL production in this study group and mortality of neonates.
Discussion
There is limited information about the diversity of the ESBL-producing genes and the clonality of the organisms involved in neonatal infections in India. It is necessary to know whether the same bacterial clones are responsible for the infections or whether dissemination of a particular ESBL-gene via genetically unrelated bacterial clones has been taking place. This study carried out in neonates highlights the presence of ESBL-producing K. pneumoniae and E. coli of diverse clonality in neonatal infections. The rate of resistance to most antibiotics was alarming, suggesting that the WHO recommended ampicillin and gentamicin combination as first line treatment of neonatal sepsis may no longer be effective31. Resistance to 3rd generation cephalosporins and monobactam was seen in these isolates, being higher amongst E. coli isolates. Ciprofloxacin resistance was more frequent in E. coli isolates, as reported previously32. Carbapenems which are reserve drugs for treatment of neonatal sepsis, showed 100 per cent sensitivity.
blaSHV genes were identified only in K. pneumoniae isolates. All different blaSHV genes identified were non-ESBL except for SHV-12 which is an ESBL type. In addition, TEM-1 was found only in K. pneumoniae and the occurrence of blaTEM-1 genes were more than blaSHV genes in K. pneumoniae isolates which is similar to a previous study on neonatal sepsis20. The E. coli isolates thus harboured a different set of β-lactamase genes in comparison to the K. pneumoniae isolates.
The majority of the blaTEM and blaSHV genes present were the classical types and were not responsible for the ESBL phenotype. It was the blaCTX-M-15 gene that was responsible for the ESBL phenotype. PFGE of the CTX-M-15-producing isolates revealed vast diversity and no epidemic clone could be seen. The clonal diversity and the possibility of dissemination of such plasmid mediated genes raise concern as experiments in the laboratory show that transfer of such genes can occur by conjugation. In addition, the presence of integron-1 in some isolates probably allows dissemination of blaCTX-M-15 gene.
Three K. pneumoniae and one E. coli isolates were found to be cefoxitin-resistant though plasmid-mediated ampC β-lactamases were not detected. This could probably occur due to loss of porins in the isolates or the presence of chromosomally-encoded ampC β-lactamases in the case of E. coli33.
Infection with ESBL-producing organisms was significantly more among the neonates for whom expressed breast milk was fed through enteral feeding tubes in comparison to the neonates fed at the breast. Previous study showed that the use of the enteral feeding tube was an independent risk factor for colonization by multiple antibiotic-resistant K. pneumoniae and E. coli but the subjects were not neonates34. Further studies with a larger population need to be done to establish this association.
Though the study is limited by the small sample size, it highlights the presence of CTX-M-15 in clonally diverse K. pneumoniae and E. coli isolates indicating that blaCTX-M-15 is probably disseminated horizontally. The high prevalence of ESBL organisms and a transmissible resistance gene (blaCTX-M) is of great concern in a country with high population density and infant mortality rate. Though carbapenems remain the most active antibiotics, the high rate of ESBL-producing organisms may compel clinicians to increase the usage of carbapenems, with a probable increase in carbapenem resistance. The spread of ESBL-producing bacteria necessitates a change towards ‘evidence-based’ treatment practices, including prudent use of antibiotics.
References
- Causes of neonatal and child mortality in India: a nationally representative mortality survey. Lancet. 2010;376:1853-60.
- [Google Scholar]
- Burden, differentials, and causes of child deaths in India. Indian J Pediatr. 2010;77:1312-21.
- [Google Scholar]
- Neonatal sepsis: a major global public health challenge. Pediatr Infect Dis J. 2009;28:S1-2.
- [Google Scholar]
- Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969-87.
- [Google Scholar]
- Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867-77.
- [Google Scholar]
- Ultrahigh resolution structure of a class A ß-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J Mol Biol. 2003;328:289-301.
- [Google Scholar]
- Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238-42.
- [Google Scholar]
- Growing group of extended-spectrum β-lacamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1-14.
- [Google Scholar]
- Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect Dis J. 2009;28:S10-8.
- [Google Scholar]
- Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from blood culture. J Med Microbiol. 2007;56:999-1000.
- [Google Scholar]
- Prevalence and clonality of extended-spectrum β-lactamases in Asia. Clin Microbiol Infect. 2008;14:159-65.
- [Google Scholar]
- Role of contaminated aspiration tubes in nosocomial outbreak of Klebsiella pneumoniae producing SHV-2 and CTX-M-15 extended-spectrum β-lactamases. J Hosp Infect. 2009;72:23-9.
- [Google Scholar]
- Extended-spectrum β-lactamase producing Klebsiella pneumoniae in neonatal intensive care unit. J Perinatol. 2008;28:685-90.
- [Google Scholar]
- Extended-spectrum β-lactamase producing Klebsiella pneumoniae from blood cultures in Puducherry, India. Indian J Med Res. 2011;134:392-5.
- [Google Scholar]
- Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332-6.
- [Google Scholar]
- Observation on integron carriage among clinical isolates of Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Indian J Med Microbiol. 2010;28:207-10.
- [Google Scholar]
- Extended spectrum beta-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian J Med Res. 2009;129:695-700.
- [Google Scholar]
- Faecal carriage of CTX-M-15-producing Klebsiella pneumoniae in patients with acute gastroenteritis. Indian J Med Res. 2009;129:599-602.
- [Google Scholar]
- Nosocomial outbreak of septicaemia in neonatal intensive care unit due to extended spectrum β-lactamase producing Klebsiella pneumoniae showing multiple mechanisms of drug resistance. Indian J Med Microbiol. 2010;28:380-4.
- [Google Scholar]
- TEM & SHV genes in extended-spectrum β-lactamase-producing Klebsiella species & their antimicrobial resistance pattern. Indian J Med Res. 2008;128:759-64.
- [Google Scholar]
- Acquisition of extended-spectrum {beta}-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal intensive care unit: molecular epidemiology and analysis of risk factors. J Med Microbiol. 2010;59:948-54.
- [Google Scholar]
- Performance standards for antimicrobial disc susceptibility tests, XVIII International Supplement (M100-S18) Wayne PA, USA: National Committee for Clinical Laboratory Standards; 2008.
- [Google Scholar]
- Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223:147-51.
- [Google Scholar]
- Occurrence of a novel SHV-type enzyme (SHV-55) among isolates of Klebsiella pneumoniae from Portuguese origin in a comparison study for extended-spectrum β-lactamase-producing evaluation. Diagn Microbiol Infect Dis. 2006;56:415-20.
- [Google Scholar]
- Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol Lett. 2002;209:161-8.
- [Google Scholar]
- Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153-62.
- [Google Scholar]
- PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41:5407-13.
- [Google Scholar]
- Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233-9.
- [Google Scholar]
- Molecular characteristics of extended spectrum β-lactamases in Eshcerichia coli and Klebsiella pneumonia and the prevalence of qnr in extended spectrum β-lactamase isolates in a tertiary care hospital in Korea. Yonsei Med J. 2010;51:768-74.
- [Google Scholar]
- Plasmid-mediated streptomycin and sulfamethoxazole resistance in Shigella flexneri 3a. Int J Antimicrob Agents. 2010;36:348-51.
- [Google Scholar]
- Antimicrobial resistance among neonatal pathogens in developing countries. Pediatr Infect Dis J. 2009;28:S19-21.
- [Google Scholar]
- Relationship between ciprofloxacin resistance and extended-spectrum beta-lactamase production in Escherichia coli and Klebsiella pneumoniae strains. Clin Microbiol Infect. 2004;10:72-5.
- [Google Scholar]
- In-vitro activity of antimicrobial agent combinations against multiresistant Acinetobacter baumannii. J Antimicrob Chemother. 1996;38:1107-8.
- [Google Scholar]
- Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999;281:517-23.
- [Google Scholar]






