Translate this page into:
Presence of multiple van genes among glycopeptide non-susceptible Staphylococcus aureus exhibiting in vitro MIC creep phenomenon: A study from north-east India
For correspondence: Dr Amitabha Bhattacharjee, Department of Microbiology, Assam University, Silchar 788 011, Assam, India e-mail: ab0404@gmail.com
-
Received: ,
Abstract
Background & objectives
The global prevalence of vancomycin-resistant Staphylococcus aureus (VRSA) has increased two fold since 2010, accounting for 2.4 per cent of S. aureus infections. The emerging hVISA isolates and their increasing trends pose a serious therapeutic challenge. The present study investigated in vitro vancomycin and teicoplanin minimum inhibitory concentration (MIC) creep in S. aureus and assessed their revertants.
Methods
A total of 845 isolates were collected for this study, and 246 were confirmed as S. aureus. Molecular characterization of vancomycin resistance was carried out by PCR assay targeting genes types viz: vanA, vanB, vanC, vanC2/C3, vanD, vanE, and vanG. MIC was determined for vancomycin and teicoplanin by agar dilution method. MIC creep and revertant analysis were done by broth dilution method in the presence and absence of antibiotics.
Results
PCR assay confirmed 12 isolates were harboured vanA, followed by vanD (n=8) and vanB (n=7). The study showed 69 isolates were screened positive for glycopeptide non-susceptibility. While analyzing vancomycin MIC creep, four isolates showed a significant increase in MIC, whereas no creep phenomenon was observed for the rest. In the case of teicoplanin, seven isolates showed the MIC creep phenomenon. Revertant analysis of all the isolates that showed MIC creep phenomenon for vancomycin and teicoplanin reverted to their original MIC when the antibiotic pressure was withdrawn.
Interpretation & conclusions
In the present study setting, glycopeptide non-susceptibility was found in eight per cent of the isolates, and the present study found the occurrence of multiple van genes from isolates calculated from a single study center will impose a serious challenge in infection control and antibiotic policy. This study also underscores that heterogenic resistant isolates, upon exposure to vancomycin and teicoplanin at a minimum level, exhibited an increase in MIC, which will impact individuals receiving glycopeptide therapy.
Keywords
Vancomycin-resistant Staphylococcus aureus (VRSA)
MIC creep
van gene
vancomycin
teicoplanin
Staphylococcus aureus is one of the predominant pathogens causing severe infections in community and hospital settings. The severity of infections leads to increased morbidity, higher healthcare costs, prolonged hospitalization, and increased risk of death1. The acquisition of resistance determinants by S. aureus has presented the greatest challenge to treating and controlling staphylococcal infections. Based on data from the Centre for Disease Control and Prevention (CDC), vancomycin-resistant Staphylococcus aureus (VRSA) is considered a serious threat to world health care2. Glycopeptide resistance is mainly due to acquiring van genes viz. vanA, vanB, vanC, vanC2/C3, vanD, vanE, vanG3. Reportedly, vanA harboring isolates exhibit a high level of inducible resistance, whereas a modest level of resistance in the case of vanB carrying isolates and low-level resistance is shown by vanC4. In a study from India5, vanA mediated vancomycin resistance was reported from a tertiary care hospital. From northeastern India, methicillin-resistant S. aureus (MRSA) associated infection was reported in 20096, where its prevalence rate was found to be 34.78 per cent. Although there is a lack of data on VRSA from this part of the country, vancomycin and vancomycin-resistant enterococci have recently been reported from a tertiary referral hospital in Assam7. Heterogeneous vancomycin-intermediate S. aureus (hVISA) exhibits vancomycin MIC within the susceptible range but has a subpopulation that confers resistance. VRSA and hVISA-associated infection pose a higher risk of treatment failure, prolonged hospital stays, higher treatment costs, and mortality. Treatment options for S.aureus-associated infections have become challenging in recent years with traditional antibiotics due to their biofilm-forming ability and acquisition of multiple resistance determinants. Besides therapeutic failure, slow clinical response and increased morbidity and relapse rate are consequences of this phenomenon8. It is also advocated that this problem should be dealt with locally with continuous evaluation of susceptibility and monitoring MIC patterns9.
S. aureus are often reported to show vancomycin MIC creep phenomenon which triggers the susceptible or intermediate S. aureus isolates towards resistance when antibiotic exposure is prolonged. MIC creep is defined as an increase in the distribution of higher vancomycin MIC values within the susceptible range10. This increasing trend of vancomycin MIC is a serious emerging threat and is reported across the globe. Many institutions have reported most MRSA strains that are susceptible to vancomycin to show a creep in the MIC of vancomycin concentration, and many authors have also reported individuals with MRSA bacteria treated with vancomycin found higher rates of clinical failure due to this phenomenon11. The current study was carried out to determine the occurrence of glycopeptide non-susceptible S. aureus in a tertiary care hospital in the northeastern part of India and in-vitro analysis of vancomycin and teicoplanin MIC creep phenomenon among the study isolates.
Material & Methods
This study was undertaken at the department of Microbiology, Silchar Medical College and Hospital, Silchar, Assam, India from September 2018 to August 2022 after obtaining the protocol approval from the Institutional Ethics Committee.
Collection of bacterial isolates
A total of 845 consecutive non-duplicate clinical isolates were collected from the individuals admitted or attended outpatient department of the Silchar Medical College. All isolates were subjected to Gram staining and cultural characteristics and were identified by VITEK® 2 compact instrument (bioMérieux, Marcy-I’Étoile, France).This sophisticated, advanced, and automated system can identify isolates efficiently within a shorter time frame. S. aureus ATCC 25923 was used as a control. The collection of isolates and their identification was done aseptically, maintaining a sterile environment.
Screening of glycopeptide non-susceptibility and minimum inhibitory concentration (MIC) determination
Screening of glycopeptides non-susceptibility was done using 6 µg/ml vancomycin and 10 µg/ml of teicoplanin (Cipla) in Brain Heart Infusion Agar (HiMedia Laboratories Pvt Ltd., Mumbai) by agar dilution method12,13. The agar dilution method has the advantage of detecting bactericidal concentration. It can rule out the presence of any subpopulation of secondary mutants that may potentially exhibit a resistance phenotype (hVISA). Isolates grown in the concentration mentioned above were suspected to be glycopeptide non-susceptible. MIC was determined for vancomycin and teicoplanin by agar dilution method according to CLSI guidelines 2017, 2020, and 202114-16. For each isolate, colonies from an overnight growth were transferred to sterile saline. The suspension was adjusted to 0.5 McFarland standards and inoculated on Muller Hinton agar (HiMedia Laboratories Pvt Ltd., Mumbai) containing 1, 2, 4, 8, 16, 32, and 64 µg/ml of vancomycin and teicoplanin individually. S. aureus ATCC 25923 was used as a negative control. Previously, laboratory-confirmed vancomycin non-susceptible S. aureus (vancomycin MIC 8 µg/ml) was taken as positive control.
Molecular detection of vancomycin resistance gene
All the screened positive isolates were further subjected to molecular characterization of vancomycin resistance by multiplex PCR targeting genes type viz: vanA, vanB, vanC, vanC2/C3, vanD, vanE, vanG. PCR was performed under the following conditions: initial denaturation at 95℃ for 3 min, denaturation at 95⁰C for 25 sec, annealing at 50⁰C for 40 sec, and extension at 72⁰C for 1min, final extension at 72⁰C for 5 min followed by 32 cycles. The primers used in the study were provided as Supplementary Table17.
In vitro MIC creep analysis
This study was done on selected eight isolates, taking two each from the MIC range of 2, 4, 8, and 16 µg/ml towards vancomycin. The isolates were subjected to serial passage in Luria Bertani broth (HiMedia Laboratories Pvt Ltd., Mumbai) containing a higher concentration of vancomycin and teicoplanin than the previous concentration. Any isolate that failed to grow in a higher antibiotic concentration was allowed to grow on the same prior concentration of vancomycin and teicoplanin. The duration of each passage was 24 h. Colonies were isolated from an overnight growth and transferred to saline for each isolate. The suspension was adjusted to 0.5 McFarland standard and inoculated in tubes containing 1, 2, 4, 8, 16, 32, 64 and 128 µg/ml of vancomycin and teicoplanin, and the process was continued for 30 days18. MIC was determined every day for each isolate for 30 days duration.
Analysis of revertant
The isolates that showed MIC creep were subjected to serial passage in LB broth at 1:1000 dilutions without antibiotic stress for 30 consecutive days. After each passage, MIC was checked against vancomycin and teicoplanin, respectively18. All the data were recorded digitally for automated instruments VITEK® 2 (bioMérieux, France) and PCR and manually for other experiments.
Results
Among 845 isolates, 246 were confirmed as S. aureus based on the VITEK® 2 compact instrument. Of the 69 were screened as glycopeptide non-susceptible. While conducting an MIC study, it was observed that 37 isolates were in the intermediate and 13 were in the resistant range against vancomycin (Table I). Towards teicoplanin, 39 isolates were within the intermediate range (16 μg/ml), and 30 were resistant (Table I). PCR assay confirmed a total of 12 isolates were harbouring vanA, followed by vanD (n=8) and vanB (n=7). Isolates with an MIC range of 2-16 μg/ml for vancomycin and 8-16 μg/ml for teicoplanin were selected for MIC creep analysis. Eight isolates were chosen for the study, covering each MIC range. While analyzing vancomycin MIC creep, four showed a significant increase in MIC (Fig. 1; Table II). In contrast, no creep phenomenon was observed for the rest of the four isolates. In the case of teicoplanin, seven isolates showed the MIC creep phenomenon (Fig. 2; Table III). While performing revertant analysis of all the isolates that showed MIC creep phenomenon for vancomycin and teicoplanin, it was observed that the isolates reverted to their initial MIC mostly in between 2-3 wk for vancomycin. The same phenomenon was observed for teicoplanin in 1-2 wk (Fig. 3 and 4; Table IV and V). However, when isolates were subjected to serial passages for 30 consecutive days without any antibiotic pressure, the MIC of vancomycin and teicoplanin came down to 1-0.25 μg/ml of the antibiotic.
| Antibiotics | Concentrations (µg/ml) | Total no. of isolates (n) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ≤2 | 2 | 4 | 8 | 16 | 32 | 64 | ≥64 | ||
| Vancomycin | 1 | 11 | 8 | 9 | 5 | 0 | 2 | 0 | 36 |
| Teicoplanin | 0 | 0 | 0 | 1 | 17 | 11 | 6 | 1 | 36 |
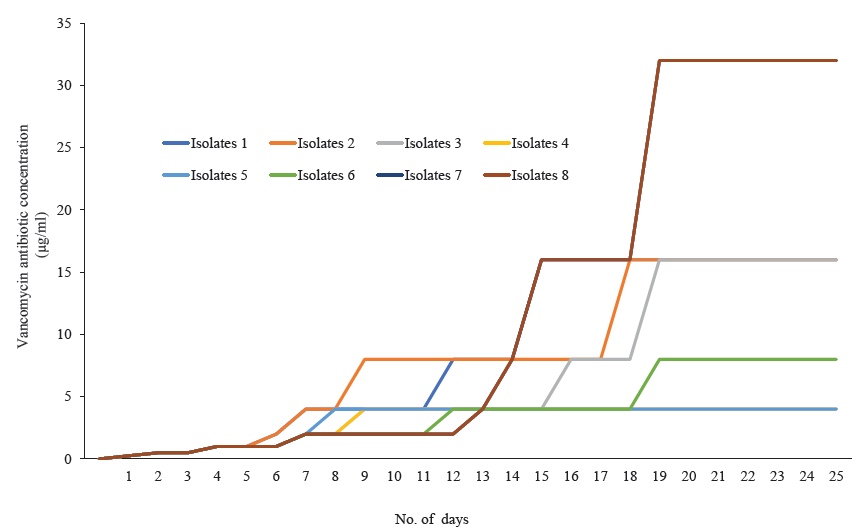
- Four Staphylococcus aureus isolates (isolates 1, 4, 6, 7) showing the minimum inhibitory concentration (MIC) creep phenomenon against vancomycin.
|
Initial MIC (µg/ml) |
Number of days and isolates grown in respective concentration of antibiotic (µg/ml) | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
| 8 | 0.25 | 0.5 | 0.5 | 1 | 1 | 2 | 4 | 4 | 4 | 4 | 4 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 2 | 4 | 4 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| 2 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 4 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 2 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| 4 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 8 | 16 | 16 | 16 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| 8 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
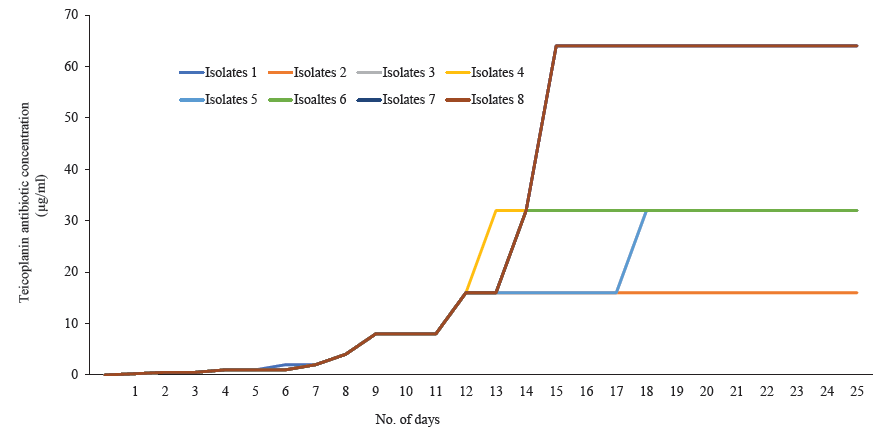
- Seven S. aureus isolates (isolates 1, 3, 4, 5, 6, 7, 8) showing the minimum inhibitory concentration (MIC) creep phenomenon against teicoplanin.
|
Initial MIC (µg/ml) |
Number of days and isolates grown in respective concentration of antibiotic (µg/ml) | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 2 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 32 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 16 | 16 | 16 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| 16 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 32 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 |
| 8 | 0.25 | 0.5 | 0.5 | 1 | 1 | 1 | 2 | 4 | 8 | 8 | 8 | 16 | 16 | 32 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 |
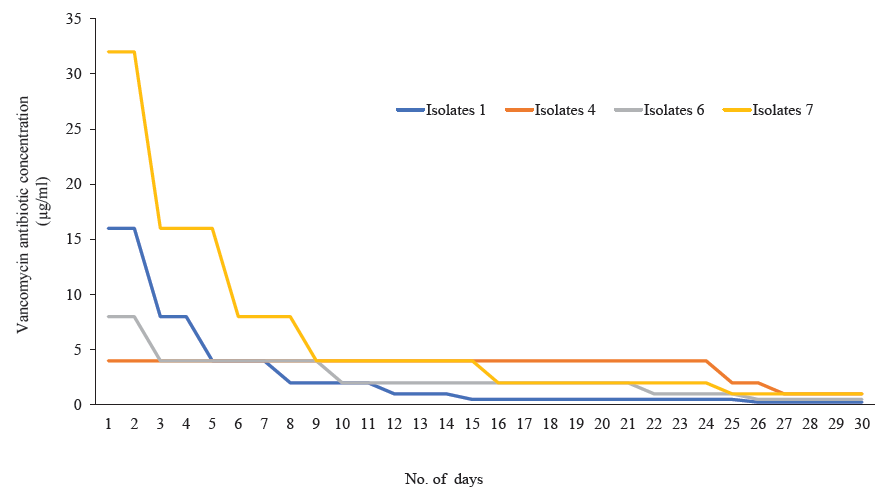
- Revertant analysis of four S. aureus isolates that showed MIC creep against vancomycin have reverted back to their respective original MIC on withdrawn of vancomycin pressure.
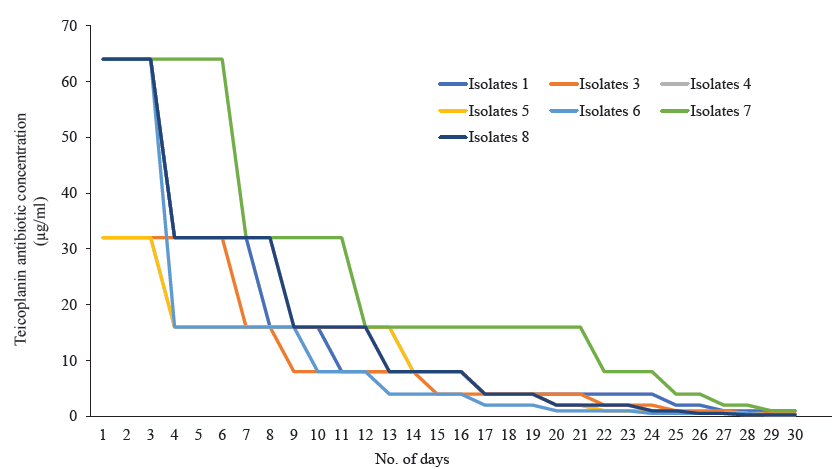
- Revertant analysis of seven S. aureus isolates that showed MIC creep against teicoplanin have reverted back to their respective original MIC on withdrawn of teicoplanin pressure.
| Isolates ID |
Initial MIC (µg/ml) |
Number of days and isolates showed growth in decreasing concentration of vancomycin (µg/ml) | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | ||
| 1 | 8 | 16 | 16 | 8 | 8 | 4 | 4 | 4 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 1 | 1 | 1 | 1 |
| 6 | 2 | 8 | 8 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| 7 | 4 | 32 | 32 | 16 | 16 | 16 | 8 | 8 | 8 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| Isolates ID | Initial MIC (µg/ml) | Number of days and isolates showed growth in decreasing concentration of teicoplanin (µg/ml) | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | ||
| 1 | 16 | 64 | 64 | 64 | 32 | 32 | 32 | 32 | 16 | 16 | 16 | 8 | 8 | 8 | 8 | 8 | 8 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 1 | 1 | 1 | 1 |
| 3 | 16 | 32 | 32 | 32 | 32 | 32 | 16 | 16 | 16 | 8 | 8 | 8 | 8 | 8 | 8 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 |
| 4 | 16 | 32 | 32 | 32 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 8 | 8 | 8 | 4 | 4 | 4 | 2 | 2 | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 5 | 16 | 32 | 32 | 32 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 8 | 8 | 8 | 8 | 4 | 4 | 4 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 6 | 16 | 32 | 32 | 32 | 16 | 16 | 16 | 16 | 16 | 16 | 8 | 8 | 8 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 7 | 16 | 64 | 64 | 64 | 64 | 64 | 64 | 32 | 32 | 32 | 32 | 32 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 8 | 8 | 8 | 4 | 4 | 2 | 2 | 1 | 1 |
| 8 | 8 | 64 | 64 | 64 | 32 | 32 | 32 | 32 | 32 | 16 | 16 | 16 | 16 | 8 | 8 | 8 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 2 | 1 | 1 | 0.5 | 0.5 | 0.25 | 0.25 | 0.25 |
Discussion
Vancomycin, the most commonly used glycopeptide antibiotic, is one of the empiric treatment options for MRSA infections19. Glycopeptide non-susceptibility among S. aureus is a serious concern that restricts treatment options within clinical settings. In the current study, glycopeptide non-susceptibility was found in eight per cent of the isolates, which agrees with a previous study where nine per cent of hVISA was recorded20. A study conducted in Italy 2012 by Tascini et al21 revealed that out of 91 clinical isolates of S. aureus, 10 (9.9%) were resistant to teicoplanin, and 5 (5.5%) were resistant to vancomycin. Shariati et al22 conducted a meta-analysis and systematic review on the prevalence of hVISA/VISA/VRSA. After data analysis from 82 studies, it was found that the overall prevalence of VRSA was 1.5 per cent, VISA was 1.7 per cent, and hVISA was 4.6 per cent, and after 2010, it increased to 2.4, 4.3, and 5.3 per cent, respectively22. In India presence of vanA was reported from S. aureus23. Where as the current study reported the presence of multiple van genes. In this study, vancomycin and teicoplanin MIC creep was observed in vitro condition that supports prolonged exposure to vancomycin increases the MIC of susceptible isolates. Furthermore 69 out of 246 S. aureus isolates were glycopeptide non-susceptible, of which 37 were of the VISA phenotype. This imposes the risk of adverse clinical outcomes if not detected early. A variable MIC range of isolates towards vancomycin and teicoplanin was observed in the study. Total 29 isolates in the susceptible MIC range against vancomycin have a potential risk of attaining hVISA phenotype in the future, severely compromising the glycopeptide treatment option. A study conducted in China showed vancomycin MIC creep in S. aureus isolates throughout the five yr study period24. There are a few contemporary reports from India where hVISA was reported within diabetic and non-diabetic individuals. They observed an occurrence rate of 6.4 per cent hVISA within MRSA isolates25. Another Indian study observed reduced vancomycin susceptibility (11.6%) in S. aureus26. Similarly, another report from south India observed 12 per cent hVISA within diverse amino acid substitution in tcaRAB, vraSR and graSR genes27. Recent studies from abroad also reported the presence of VISA/VRSA among MRSA strains28. A study from Saudi Arabia29 has reported vancomycin MIC creep in S. aureus over three years, which declined subsequently over the next three years’ time. VISA/hVISA phenotypes are reported to be associated with a mutation on vraSR and graSR, two-component regulation systems. It was observed that the constructs (mutants) demonstrated a remarkable increase in vancomycin MIC30. Thus, the in vitro gradual increase in vancomycin MIC in the current study might have a link with mutations in regulatory regions. A study from the USA31 highlighted the MIC creep phenomenon over some time towards vancomycin. Recently, a study from India showed progressive MIC creep towards teicoplanin32. In the present study, all the isolates that showed MIC creep against vancomycin and teicoplanin reverted to a susceptible MIC range when antibiotic stress was withdrawn for 30 days. However, no study could be found to compare with the findings of the present study. Yeh et al33 in 2012 showed increased vancomycin usage, resulting in vancomycin MIC creep in MRSA. It was also reported that vancomycin usage in 30 days before the isolation of a S. aureus culture had a higher MIC. However, this increase could not be correlated with higher mortality33. The findings of this study will augment global knowledge of antimicrobial resistance. Our observations on low vancomycin MIC (4 µg/ml) of vanD harboring isolates and moderate to high vancomycin MIC (8-64 µg/ml) of vanA and vanB harboring isolates could underscore how these resistance determinants confer different phenotypes. In the present study, the in vitro MIC creep phenomenon was observed in laboratory conditions, advocating studies to be undertaken over two to three yr to understand and correlate with glycopeptide usage and any increase in MIC. The findings of this study emphasize the need for a local epidemiological cut-off point for screening resistant pathogens. This warrants designing future diagnostics that can effectively detect heterogeneous resistant populations of bacteria. The current study could predict how these glycopeptide nonsusceptible isolates can attain a higher inhibitory concentration within the patient population when initiating glycopeptide treatment. This also highlights the adoption of testing facilities for glycopeptide non-susceptibility within routine microbiology laboratories. However, the information of the current investigation is restricted to in vitro analysis only.
Overall, the study found the prevalence of multiple van genes within a single study centre, which poses a severe challenge to treatment options. The presence of van genes among clinical isolates of S. aureus is a serious concern as the hospital environment acts as a reservoir for the resistance determinants. The emergence of hVISA is a significant threat that requires urgent screening and proper reporting.The present study is retrospective, and this advocates further need for prospective investigation under the umbrella of an antibiotic stewardship programme, thereby adopting control measures to contain this spread and detect resistant strains. This study also signifies that the heterogenic resistance strains upon exposure to vancomycin and teicoplanin at a minimum level could increase MIC significantly, particularly in individuals receiving therapeutic intervention in real time.
Financial support & sponsorship
This study received financial support from the Council of Scientific and Industrial Research, Government of India (No. 27(0321)/17/EMR-II).
Conflicts of Interest
None.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that no artificial intelligence (AI)-assisted technology was used to assist in the writing of the manuscript, and no images were manipulated using AI.
References
- Evaluation of vancomycin MIC creep in methicillin-resistant Staphylococcus aureus infections – a systematic review and meta-analysis. Clin Microbial Infect. 2018;24:97-104.
- [Google Scholar]
- Comparison of phenotypic versus genotypic methods in the detection of methicillin resistance in Staphylococcus aureus. Indian J Med Res. 2008;127:78-84.
- [PubMed] [Google Scholar]
- Glycopeptide antibiotic resistance. Annu Rev Pharmacol Toxicol. 2002;42:381.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686-707.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Emergence of vanA gene among vancomycin-resistant enterococci in a tertiary care hospital of North-East India. Indian J Med Res. 2016;143:357-61.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus in Assam. Indian J Crit Care Med. 2009;13:156.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Trends in antimicrobial resistance in a tertiary care hospital of Assam, India. J Pure Appl Microbiol. 2023;17:1591-604.
- [CrossRef] [Google Scholar]
- Treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in children: a reappraisal of vancomycin. Curr Infect Dis Rep. 2019;21:1-8.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin MIC creep in MRSA blood culture isolates from germany: A regional problem? Eur J Clin Microbiol Infect Dis. 2011;30:677-83.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin, teicoplanin, daptomycin, and linezolid MIC creep in methicillin-resistant Staphylococcus aureus is associated with clonality. Medicine (Baltimore). 2016;95:e5060.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Vancomycin MICs do not predict the outcome of methicillin-resistant Staphylococcus aureus bloodstream infections in correctly treated patients. J Antimicrob Chemother. 2012;67:1760-8.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin MICs did not creep in Staphylococcus aureus isolates from 2002 to 2006 in a setting with low vancomycin usage. J Antimicrob Chemother. 2008;62:773-5.
- [CrossRef] [PubMed] [Google Scholar]
- Investigation of reduced susceptibility to glycopeptides among methicillin-resistant Staphylococcus aureus isolates from patients in Ireland and evaluation of agar screening methods for detection of heterogeneously glycopeptide-intermediate S. aureus. J Clin Microbiol. 2007;45:3263-9.
- [Google Scholar]
- M100. Performance standards for antimicrobial susceptibility testing 27th edition; 2017. Available from: https://clsi.org/media/1469/m100s27_sample.pdf, accessed on October 15, 2022.
- M100. Performance standards for antimicrobial susceptibility testing 30th edition; 2020. Available from: https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf, accessed on October 15, 2022.
- M100. Performance standards for antimicrobial susceptibility testing 31st edition; 2021. Available from: https://clsi.org/media/z2uhcbmv/m100ed31_sample.pdf, accessed on October 15, 2022.
- Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol. 2004;42:5857-60.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Escherichia coli encoding blaNDM-5 associated with community-acquired urinary tract infections with unusual MIC creep-like phenomenon against imipenem. J Glob Antimicrob Resist. 2018;14:228-32.
- [CrossRef] [PubMed] [Google Scholar]
- Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10:12689.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Vancomycin MIC distribution among methicillin-resistant Staphylococcus aureus. Is reduced vancomycin susceptibility related to MIC creep? Open Access Maced J Med Sci. 2019;7:12.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Comparison of teicoplanin and vancomycin in vitro activity on clinical isolates of Staphylococcus aureus. J Chemother.. 2012;24:187-90.
- [CrossRef] [PubMed] [Google Scholar]
- Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: A systematic review and meta-analysis. Sci Rep. 2020;10:12689.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Molecular confirmation of vancomycin-resistant Staphylococcus aureus with vanA gene from a hospital in kathmandu. Int J Microbiol. 2021;2021:3847347.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Vancomycin MIC creep in methicillin-resistant Staphylococcus aureus (MRSA) isolates from 2006 to 2010 in a hospital in China. Indian J Med Microbiol. 2015;33:262-6.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular and clinical features of heterogeneous vancomycin-intermediate Staphylococcus aureus in tertiary care hospitals in South India. Sultan Qaboos Univ Med J. 2023;23:447.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Recent pattern of antibiotic resistance in Staphylococcus aureus clinical isolates in Eastern India and the emergence of reduced susceptibility to vancomycin. J Lab Physicians. 2019;11:340-5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Genomic insights on heterogeneous resistance to vancomycin and teicoplanin in Methicillin-resistant Staphylococcus aureus: A first report from South India. PLoS One. 2019;14:e0227009.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Detection of vancomycin resistance among hospital and community-acquired methicillin-resistant Staphylococcus aureus isolates. Egypt J Med Microbiol. 2023;32:45-52.
- [CrossRef] [Google Scholar]
- Occurrence of vancomycin MIC creep in methicillin resistant isolates in Saudi Arabia. J Infect Public Health. 2020;13:1576-9.
- [CrossRef] [PubMed] [Google Scholar]
- Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:1231-4.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–05. J Antimicrob Chemother. 2007;60:788-94.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus at an Indian tertiary hospital: Antimicrobial susceptibility and minimum inhibitory concentration (MIC) creep of antimicrobial agents. J Glob Antimicrob Resist. 2019;17:98-102.
- [CrossRef] [PubMed] [Google Scholar]
- Impact of vancomycin MIC creep on patients with methicillin-resistant Staphylococcus aureus bacteremia. J Microbiol Immunol Infect. 2012;45:214-20.
- [CrossRef] [PubMed] [Google Scholar]






